
Applying Localized Feature Selection in Python and MQL5
Introduction
In financial market analysis, indicators often exhibit varying effectiveness as underlying conditions change. For example, fluctuating volatility can render previously reliable indicators ineffective as market regimes shift. This variability explains the proliferation of indicators used by traders, as no single indicator can consistently perform well across all market conditions. From a machine learning perspective, this calls for a flexible feature selection technique that can accommodate such dynamic behavior.
Many common feature selection algorithms prioritize features that show predictive power across the entire feature space. These features are often favored even when their relationships with the target variable are nonlinear or influenced by other features. However, this global bias can be problematic, as modern nonlinear models can extract valuable insights from features with strong local predictive capabilities or whose relationships with the target variable shift within specific regions of the feature space.
In this article, we explore a feature selection algorithm introduced in the paper 'Local Feature Selection for Data Classification' by Narges Armanfard, James P. Reilly, and Majid Komeili. This method aims to identify predictive features that are often overlooked by traditional selection techniques due to their limited global utility. We will begin with a general overview of the algorithm, followed by its implementation in Python to create classifier models suitable for export to MetaTrader 5.
Local feature selection
Successful machine learning relies on selecting informative features that contribute to solving the problem. In supervised classification, features should effectively differentiate between data categories. However, identifying these informative features can be challenging, as uninformative ones can introduce noise and degrade model performance. As a result, feature selection is often a critical initial step in building predictive models.
Unlike traditional methods that seek a single optimal feature subset for all data, Local Feature Selection (LFS) identifies optimal subsets for specific local regions. This adaptability could be particularly useful for handling non-stationary data. Moreover, LFS incorporates a classifier that accounts for the varying feature subsets used across different samples. It achieves this through class-wise clustering, selecting features that minimize intra-class distances while maximizing inter-class distances.
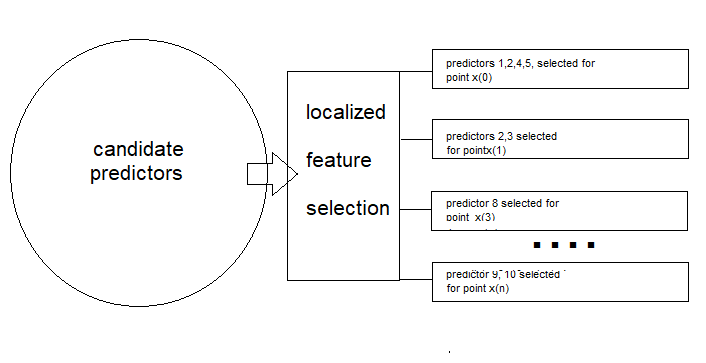
This approach identifies a locally optimal feature subspace in overlapping regions, ensuring that each sample is represented in multiple feature spaces. To better understand the concept, consider a scenario where a telecommunications company aims to predict customer churn—identifying customers likely to close their accounts. The company collects various customer characteristics, including:
- Customer tenure: How long has the customer been with the company?
- Monthly bill: How much does the customer pay each month?
- Customer's weight and height.
- Number of calls made to customer service: How often does the customer contact support.
Imagine selecting two loyal customers who have been with the company for many years. For each of the features described, there would likely be minimal differences between these loyal customers, as they belong to the same class. Now, contrast this with the difference between a long-time customer and one who canceled their subscription shortly after signing up. While their weight and height may not differ much, other relevant predictors would likely show significant variation.
The loyal customer would obviously have a much longer tenure, might be more willing to opt for a higher-priced subscription package, and is more likely to contact customer support when issues arise rather than canceling in frustration. Meanwhile, metrics like weight and height would remain close to the population average and wouldn't contribute significantly to distinguishing these customer types.
Analyzing individual feature values in pairs using Euclidean distance reveals that the most relevant predictors will have the greatest inter-class distance between customers, while the least relevant predictors will exhibit the smallest inter-class distance. This makes the selection of effective predictors clear: we prioritize pairs with low intra-class distance and high inter-class distance.
While this approach seems effective, it falls short in accounting for local variations within the data. To address this, we must consider how predictive power can differ across various feature domains. Imagine a dataset with two classes, where one class is divided into two distinct subsets. A scatter plot of two features from this dataset illustrates that the first subset may be well-separated from Class 1 using the x1 variable, but not x2. Conversely, the second subset may be well-separated using x2, but not x1.
If we only consider inter-class separation, the algorithm might mistakenly select both x1 and x2, even though only one is truly effective in each subset. This happens because the algorithm could prioritize the overall large distance between the two subsets over the smaller, more relevant distances within each subset. To solve this, the authors of the cited paper introduced a weighting scheme for the distances. By assigning higher weights to pairs of cases that are closer together and lower weights to pairs that are farther apart, the algorithm can reduce the influence of outliers within a class. This considers both class memberships and the global distribution of distances.
In summary, the LFS algorithm, as described in the cited paper, consists of two main components. The first is the feature selection process, where a subset of features is selected for each sample. The second component involves a localized mechanism that measures the similarity of a test sample to a specific class, which is used for inference purposes.
Feature selection
In this section, we will describe the learning procedure employed by the LFS method, step by step, with a little math. We begin with the expected structure of the training data. The implementation of localized feature selection is performed on a dataset with N training samples, classified into Z class labels, and accompanied by M features or predictor candidates.
The training data can be represented as a matrix X, where the rows correspond to the samples and the columns represent distinct predictor candidates. Thus, matrix X has N rows and M columns. Each sample is denoted as X(i), referring to the i-th row in the matrix. The class labels are stored in a separate column vector Y, with each label mapped to a corresponding sample (row) in the matrix.
The ultimate goal of applying the LFS method is to determine, for each training sample X(i), an M-sized binary vector F(i) that indicates which candidate predictors are most relevant for determining the corresponding class label. The matrix F will have the same dimensions as X.
Using Euclidean distance, the algorithm aims to minimize the average distance between the current sample and other samples with the same class label, while maximizing the average distance between the current sample and those with different class labels. Additionally, the distances must be weighted to favor samples in the same neighborhood as the current sample, introducing the weights column vector W. Since the weights (W) and the binary vector F(i) are not initially available, an iterative procedure is used to estimate both the optimal W and F(i) vectors.
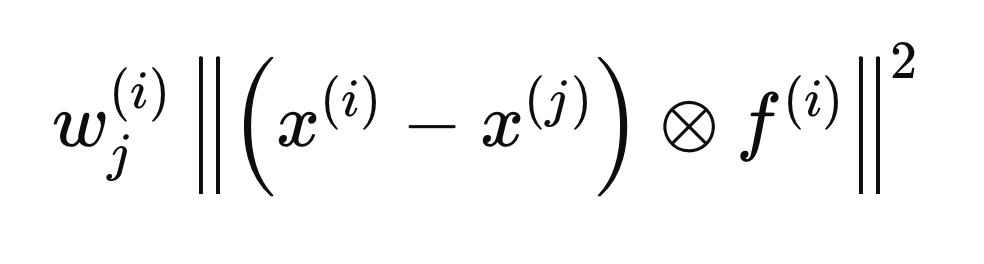
Computing intra-class and inter-class distances
Each step described in the following sections pertains to calculations performed for a single sample, X(i), to determine the optimal F(i) vector. The process begins by initializing all entries of F to zero and setting the initial weights to 1. Next, we calculate the intra-class and inter-class distances regarding X(i). The inclusion of the F(i) vector in the distance calculations ensures that only the variables deemed relevant (those equal to 1) are considered. For mathematical convenience, the Euclidean distances are squared, leading to the following distance equation.
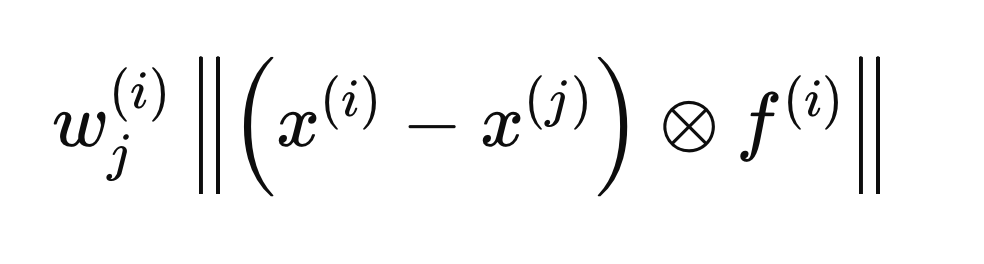
The circle with an enclosed "x" denotes an operator for element-wise multiplication. The intra-class and inter-class distances are computed using the formula above, but with different j elements (rows) of X. The intra-class distance is calculated using the j elements that share the same class label as X(i),
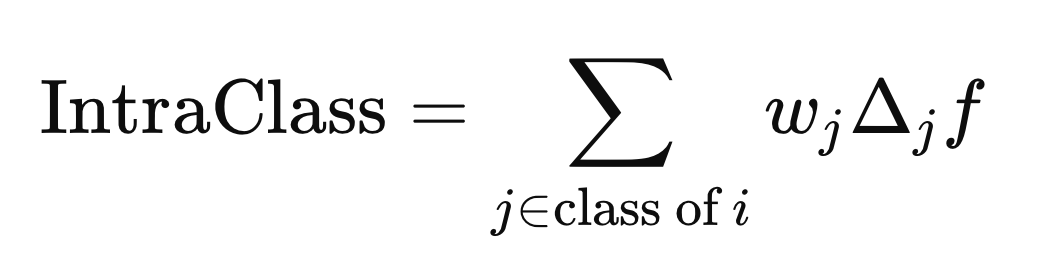
while the inter-class distance is computed using the j elements with any class label different from Y(i).
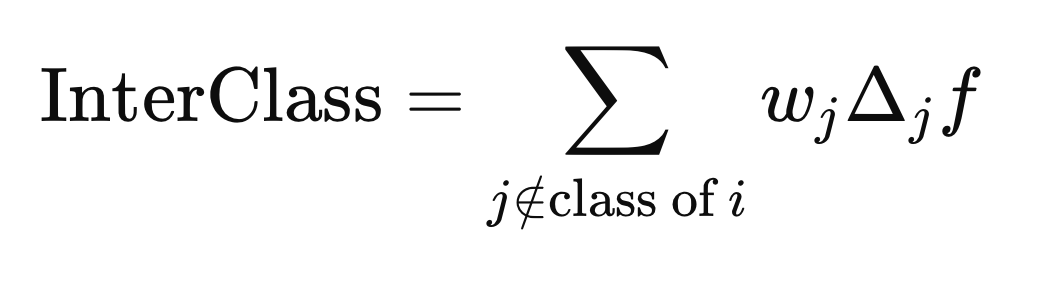
Calculating the weights
For sample X(i), we calculate a vector of weights (W), which is N-long, such that if X(j) is far from X(i), its weight should be small, and conversely, if it is nearby, the weight should be larger. The weighting should not penalize samples simply because they have a different class label. Since F(i) is not yet optimal, the variables selected to define the basis of neighborhoods are still unknown. The cited paper addresses this issue by averaging the weights calculated from previous iterations of weight refinement.
When an F vector is included in defining the distance between two samples, it is considered within the metric space defined by F(i). The calculation of optimal weights is performed by defining distances in terms of a different metric space, which we will refer to as F(z), as given by the formula below.
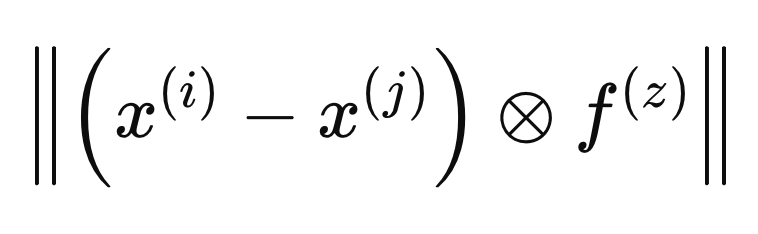
To ensure that the weights do not penalize samples simply for being in a different class, we calculate the minimum distance between X(i) and all other samples of the same class in the metric space defined by F(z).
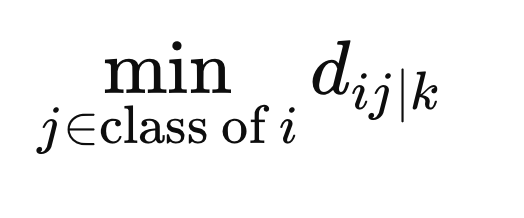
Additionally, we compute the minimum distance from samples with a different class label to X(i).
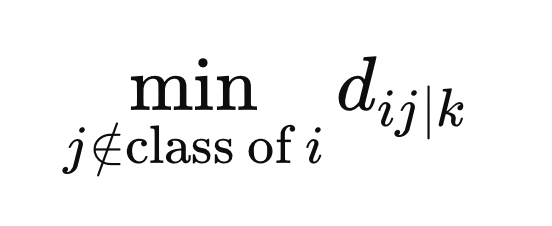
These are the final values needed to define the weights. The weights are calculated as the average across all metric spaces, given by the negative exponential of the difference between the distance and the minimum distance for a particular metric space, z.
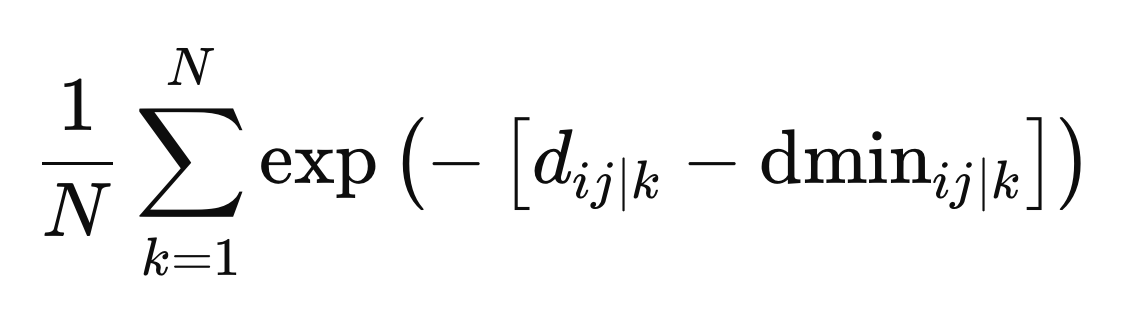
Conflicting objectives
At this stage, we have obtained the optimal weights, allowing us to address the challenge of finding the right balance between inter-class and intra-class separation. This involves reconciling two conflicting objectives: minimizing intra-class separation (making data points within the same class as similar as possible) and maximizing inter-class separation (making different classes as distinct as possible). Achieving both objectives perfectly with the same set of predictors is usually infeasible.
A viable approach is the Epsilon-Constraint Method, which finds a compromise between these conflicting goals. This method works by first solving one of the optimization problems (usually the maximization problem), and then addressing the minimization problem with the added constraint that the maximized function remains above a certain threshold.
First, we maximize the inter-class separation and record the maximum value of this function, denoted as epsilon (ϵ), which represents the highest possible inter-class separation. Next, we minimize intra-class separation for various values of a parameter β (ranging from 0 to 1), with the constraint that the inter-class separation for the minimized solution must remain greater than or equal to βϵ.
The parameter β serves as a compromise factor, balancing the focus between the two objectives: when β is set to 1, inter-class separation takes full priority, while when β is set to 0, the focus shifts entirely to minimizing intra-class separation. Four constraints are imposed on both optimization tasks:
- All elements of F must be between 0 and 1, inclusive.
- The sum of the elements of an F vector must be less than or equal to a user-specified hyperparameter, which governs the maximum number of predictors that can be activated.
- The sum of the elements of an F vector must be greater than or equal to one, ensuring that at least one predictor is activated for each sample.
For intra-class minimization, there is an additional constraint inherited from the initial maximization operation: the value of the function maximization must be at least equal to the product of β and ϵ.
The functions and constraints involved are linear, indicating that the optimization tasks are linear programming problems. Standard linear programming problems aim to maximize an objective function subject to constraints that specify thresholds that must not be exceeded.
Linear programming involves optimizing a linear objective function subject to linear constraints. The objective function, typically denoted as "z," is a linear combination of decision variables. Constraints are expressed as linear inequalities or equalities, limiting the values of the decision variables. Beyond the user-specified constraints, there are implicit non-negativity constraints on the decision variables and non-negativity constraints on the right-hand sides of the inequalities.
While the standard form assumes non-negative decision variables and "less-than-or-equal" inequalities, these restrictions can be relaxed through transformations. By multiplying both sides of an inequality by -1, we can handle "greater-than-or-equal" inequalities and negative right-hand sides. Additionally, non-positive coefficients involving decision variables can be transformed into positive coefficients by creating new variables.
The interior-point method is an efficient algorithm for solving linear programming problems, especially when dealing with large-scale optimization tasks. Our Python implementation will employ this method to efficiently find an optimal solution. Once convergence is reached, we will obtain an optimal F(i) vector. However, it is important to note that these values are not in the required format (either 1s or 0s). This is corrected in the final step of the LFS method.
Beta trials
The problem with the calculated F(i) vector is that it consists of real values rather than binary values. The goal of the LFS procedure is to identify the most relevant variables for each sample, which is represented by a binary F matrix where values are either 0 or 1. A value of 0 indicates that the corresponding variable is deemed irrelevant or skipped.
To convert the real values of the F(i) vector into binary values, we use a Monte-Carlo method to find the best binary equivalent. This involves repeating the process a user-specified number of times, which is a key hyperparameter of the LFS method. For each iteration, we start with a binary vector where each predictor candidate is initially set to 1, using the continuous F(i) values as probabilities for each predictor. We then check if the binary vector satisfies the constraints of the minimization procedure and calculate its objective function value. The binary vector with the minimum objective function value is chosen as the final F(i) vector.
Post-processing for feature selection
LFS independently selects optimal predictor candidates for each sample, making it impractical to report a single definitive set. To address this, we count the frequency of each predictor's inclusion in optimal subsets. This allows users to set a threshold and identify the most frequently appearing predictors as the most relevant. Importantly, the relevance of a predictor within this set does not imply its individual worth; its value might lie in its interaction with other predictors.
This is a key advantage of LFS: its ability to pinpoint predictors that might be individually insignificant but valuable when combined with others. This preprocessing step is important for modern prediction models, which excel at discerning complex relationships between variables. By eliminating irrelevant predictors, LFS streamlines the modeling process and enhances the model performance.
Python implementation: LFSpy
In this section, we explore the practical application of the LFS algorithm, focusing first on its use as a feature selection technique and briefly discussing its data classification capabilities. All demonstrations will be conducted in Python using the LFSpy package, which implements both the feature selection and data classification aspects of the LFS algorithm. The package is available on PyPI, where detailed information about it can be found.
First, install the LFSpy package.
pip install LFSpy
Next, import the LocalFeatureSelection class from LFSpy.
from LFSpy import LocalFeatureSelection
An instance of LocalFeatureSelection can be created by calling the parametric constructor.
lfs = LocalFeatureSelection(alpha=8,tau=2,n_beta=20,nrrp=2000)
The constructor supports the following optional parameters:
| Parameter Name | Data Type | Description |
|---|---|---|
| alpha | integer | The maximum number of selected predictors out of all predictor candidates. The default value is 19. |
| gamma | double | A tolerance level governing the ratio of samples with differing class labels to those with the same class label within a local region. The default value is 0.2. |
| tau | integer | The number of iterations through the entire dataset (equivalent to the number of epochs in traditional machine learning). The default is 2, and it's recommended to set this value to a single-digit number, typically no more than 5. |
| sigma | double | Controls the weighting of observations based on their distance. A value greater than 1 reduces the weighting. The default is 1. |
| n_beta | integer | The number of beta values tested when converting the continuous F vectors to their binary equivalents. |
| nrrp | integer | The number of iterations for beta trials. This value should be at least 500, increasing with the size of the training dataset. The default is 2000. |
| knn | integer | Applies specifically to classification tasks. It specifies the number of nearest neighbors to compare for categorization. The default value is 1. |
After initializing an instance of the LFSpy class, we use the fit() method with at least two input parameters: a two-dimensional matrix of training samples, consisting of candidate predictors, and a one-dimensional array of corresponding class labels.
lfs.fit(xtrain,ytrain)
Once the model is fitted, calling fstar returns the F inclusion matrix, which consists of ones and zeros to indicate the selected features. Note that this matrix is transposed relative to the orientation of the training samples.
fstar = lfs.fstar
The predict() method is used to classify test samples based on the learned model and returns the class labels corresponding to the test data.
predicted_classes = lfs.predict(test_samples)
The score() method calculates the model’s accuracy by comparing the predicted class labels with the known labels. It returns the fraction of test samples that were correctly classified.
accuracy = lfs.score(test_data,test_labels)
Examples of LFSpy
For the first practical demonstration, we generate several thousand uniformly distributed random variables within the interval [−1,1][−1,1]. These variables are arranged into a matrix with a specified number of columns. We then create a vector of {0, 1} labels corresponding to each row, depending on whether the values in two arbitrary columns are both negative or both positive. The goal of this demonstration is to determine whether the LFS method can identify the most relevant predictors in this dataset. We evaluate the results by summing the number of times each predictor is selected (indicated by a 1) in the F binary inclusion matrix. The code implementing this test is shown below.
import numpy as np import pandas as pd from LFSpy import LocalFeatureSelection from timeit import default_timer as timer #number of random numbers to generate datalen = 500 #number of features the dataset will have datavars = 5 #set random number seed rng_seed = 125 rng = np.random.default_rng(rng_seed) #generate the numbers data = rng.uniform(-1.0,1.0,size=datalen) #shape our dataset data = data.reshape([datalen//datavars,datavars]) #set up container for class labels class_labels = np.zeros(shape=data.shape[0],dtype=np.uint8) #set the class labels for i in range(data.shape[0]): class_labels[i] = 1 if (data[i,1] > 0.0 and data[i,2] > 0.0) or (data[i,1] < 0.0 and data[i,2] < 0.0) else 0 #partition our training data xtrain = data ytrain = class_labels #initialize the LFS object lfs = LocalFeatureSelection(rr_seed=rng_seed,alpha=8,tau=2,n_beta=20,nrrp=2000) #start timer start = timer() #train the model lfs.fit(xtrain,ytrain) #output training duration print("Training done in ", timer()-start , " seconds. ") #get the inclusion matrix fstar = lfs.fstar #add up all ones for each row of the inclusion matrix ibins = fstar.sum(axis=1) #calculate the percent of times a candidate was selected original_crits = 100.0 * ibins.astype(np.float64)/np.float64(ytrain.shape[0]) #output the results print("------------------------------> Percent of times selected <------------------------------" ) for i in range(original_crits.shape[0]): print( f" Variable at column {i}, selected {original_crits[i]} %")
The output from running LFSdemo.py
Training done in 45.84896759999992 seconds. Python ------------------------------> Percent of times selected <------------------------------ Python Variable at column 0, selected 19.0 % Python Variable at column 1, selected 81.0 % Python Variable at column 2, selected 87.0 % Python Variable at column 3, selected 20.0 % Python Variable at column 4, selected 18.0 %
It is intriguing that one of the relevant variables was selected slightly more frequently than the other, despite their identical roles in predicting the class. This suggests that subtle nuances within the data might be influencing the selection process. What is clear is that both variables were consistently chosen more often than irrelevant predictors, indicating their significance in determining the class. The algorithm's relatively slow execution is likely due to its single-threaded nature, potentially hindering its performance on larger datasets.
LFS for data classification
Given LFS's local nature, constructing a classifier from it requires more effort compared to traditional, globally biased feature selection methods. The referenced paper discusses a proposed classifier architecture, which we will not delve into here. Interested readers are encouraged to refer to the cited paper for full details. In this section, we will focus on the implementation.
The predict() method of the LocalFeatureSelection class assesses class similarity. It takes test data that matches the structure of the training data and returns predicted class labels based on the patterns learned by the trained LFS model. In the next code demonstration, we will extend the previous script to build an LFS classifier model, export it in JSON format, load it using an MQL5 script, and classify an out-of-sample dataset. The code used to export an LFS model is contained in JsonModel.py. This file defines the lfspy2json() function, which serializes the state and parameters of a LocalFeatureSelection model into a JSON file. This allows the model to be saved in a format that can be easily read and used in MQL5 code, facilitating integration with MetaTrader 5. The full code is shown below.
# Copyright 2024, MetaQuotes Ltd. # https://www.mql5.com from LFSpy import LocalFeatureSelection import json MQL5_FILES_FOLDER = "MQL5\\FILES" MQL5_COMMON_FOLDER = "FILES" def lfspy2json(lfs_model:LocalFeatureSelection, filename:str): """ function export a LFSpy model to json format readable from MQL5 code. param: lfs_model should be an instance of LocalFeatureSelection param: filename or path to file where lfs_model parameters will be written to """ if not isinstance(lfs_model,LocalFeatureSelection): raise TypeError(f'invalid type supplied, "lfs_model" should be an instance of LocalFeatureSelection') if len(filename) < 1 or not isinstance(filename,str): raise TypeError(f'invalid filename supplied') jm = { "alpha":lfs_model.alpha, "gamma":lfs_model.gamma, "tau":lfs_model.tau, "sigma":lfs_model.sigma, "n_beta":lfs_model.n_beta, "nrrp":lfs_model.nrrp, "knn":lfs_model.knn, "rr_seed":lfs_model.rr_seed, "num_observations":lfs_model.training_data.shape[1], "num_features":lfs_model.training_data.shape[0], "training_data":lfs_model.training_data.tolist(), "training_labels":lfs_model.training_labels.tolist(), "fstar":lfs_model.fstar.tolist() } with open(filename,'w') as file: json.dump(jm,file,indent=None,separators=(',', ':')) return
The function takes a LocalFeatureSelection object and a file name as inputs. It serializes the model parameters as a JSON object and saves it under the specified file name. The module also defines two constants, MQL5_FILES_FOLDER and MQL5_COMMON_FOLDER, which represent the directory paths for accessible folders in a standard MetaTrader 5 installation. This is only one part of the solution for integrating with MetaTrader 5. The other part is implemented in MQL5 code, which is presented in lfspy.mqh. This included file contains the definition of the Clfspy class, which facilitates loading an LFS model saved in JSON format for inference purposes. The full code is provided below.
//+------------------------------------------------------------------+ //| lfspy.mqh | //| Copyright 2024, MetaQuotes Ltd. | //| https://www.mql5.com | //+------------------------------------------------------------------+ #property copyright "Copyright 2024, MetaQuotes Ltd." #property link "https://www.mql5.com" #include<JAson.mqh> #include<Files/FileTxt.mqh> #include<np.mqh> //+------------------------------------------------------------------+ //|structure of model parameters | //+------------------------------------------------------------------+ struct LFS_PARAMS { int alpha; int tau; int n_beta; int nrrp; int knn; int rr_seed; int sigma; ulong num_features; double gamma; }; //+------------------------------------------------------------------+ //| class encapsulates LFSpy model | //+------------------------------------------------------------------+ class Clfspy { private: bool loaded; LFS_PARAMS model_params; matrix train_data, fstar; vector train_labels; //+------------------------------------------------------------------+ //| helper function for parsing model from file | //+------------------------------------------------------------------+ bool fromJSON(CJAVal &jsonmodel) { model_params.alpha = (int)jsonmodel["alpha"].ToInt(); model_params.tau = (int)jsonmodel["tau"].ToInt(); model_params.sigma = (int)jsonmodel["sigma"].ToInt(); model_params.n_beta = (int)jsonmodel["n_beta"].ToInt(); model_params.nrrp = (int)jsonmodel["nrrp"].ToInt(); model_params.knn = (int)jsonmodel["knn"].ToInt(); model_params.rr_seed = (int)jsonmodel["rr_seed"].ToInt(); model_params.gamma = jsonmodel["gamma"].ToDbl(); ulong observations = (ulong)jsonmodel["num_observations"].ToInt(); model_params.num_features = (ulong)jsonmodel["num_features"].ToInt(); if(!train_data.Resize(model_params.num_features,observations) || !train_labels.Resize(observations) || !fstar.Resize(model_params.num_features,observations)) { Print(__FUNCTION__, " error ", GetLastError()); return false; } for(int i=0; i<int(model_params.num_features); i++) { for(int j = 0; j<int(observations); j++) { if(i==0) train_labels[j] = jsonmodel["training_labels"][j].ToDbl(); train_data[i][j] = jsonmodel["training_data"][i][j].ToDbl(); fstar[i][j] = jsonmodel["fstar"][i][j].ToDbl(); } } return true; } //+------------------------------------------------------------------+ //| helper classification function | //+------------------------------------------------------------------+ matrix classification(matrix &testing_data) { int N = int(train_labels.Size()); int H = int(testing_data.Cols()); matrix out(H,2); for(int i = 0; i<H; i++) { vector column = testing_data.Col(i); vector result = class_sim(column,train_data,train_labels,fstar,model_params.gamma,model_params.knn); if(!out.Row(result,i)) { Print(__FUNCTION__, " row insertion failure ", GetLastError()); return matrix::Zeros(1,1); } } return out; } //+------------------------------------------------------------------+ //| internal feature classification function | //+------------------------------------------------------------------+ vector class_sim(vector &test,matrix &patterns,vector& targets, matrix &f_star, double gamma, int knn) { int N = int(targets.Size()); int n_nt_cls_1 = (int)targets.Sum(); int n_nt_cls_2 = N - n_nt_cls_1; int M = int(patterns.Rows()); int NC1 = 0; int NC2 = 0; vector S = vector::Zeros(N); S.Fill(double("inf")); vector NoNNC1knn = vector::Zeros(N); vector NoNNC2knn = vector::Zeros(N); vector NoNNC1 = vector::Zeros(N); vector NoNNC2 = vector::Zeros(N); vector radious = vector::Zeros(N); double r = 0; int k = 0; for(int i = 0; i<N; i++) { vector fs = f_star.Col(i); matrix xpatterns = patterns * np::repeat_vector_as_rows_cols(fs,patterns.Cols(),false); vector testpr = test * fs; vector mtestpr = (-1.0 * testpr); matrix testprmat = np::repeat_vector_as_rows_cols(mtestpr,xpatterns.Cols(),false); vector dist = MathAbs(sqrt((pow(testprmat + xpatterns,2.0)).Sum(0))); vector min1 = dist; np::sort(min1); vector min_uniq = np::unique(min1); int m = -1; int no_nereser = 0; vector NN(dist.Size()); while(no_nereser<int(knn)) { m+=1; double a1 = min_uniq[m]; for(ulong j = 0; j<dist.Size(); j++) NN[j]=(dist[j]<=a1)?1.0:0.0; no_nereser = (int)NN.Sum(); } vector bitNN = np::bitwiseAnd(NN,targets); vector Not = np::bitwiseNot(targets); NoNNC1knn[i] = bitNN.Sum(); bitNN = np::bitwiseAnd(NN,Not); NoNNC2knn[i] = bitNN.Sum(); vector A(fs.Size()); for(ulong v =0; v<A.Size(); v++) A[v] = (fs[v]==0.0)?1.0:0.0; vector f1(patterns.Cols()); vector f2(patterns.Cols()); if(A.Sum()<double(M)) { for(ulong v =0; v<A.Size(); v++) A[v] = (A[v]==1.0)?0.0:1.0; matrix amask = matrix::Ones(patterns.Rows(), patterns.Cols()); amask *= np::repeat_vector_as_rows_cols(A,patterns.Cols(),false); matrix patternsp = patterns*amask; vector testp = test*(amask.Col(0)); vector testa = patternsp.Col(i) - testp; vector col = patternsp.Col(i); matrix colmat = np::repeat_vector_as_rows_cols(col,patternsp.Cols(),false); double Dist_test = MathAbs(sqrt((pow(col - testp,2.0)).Sum())); vector Dist_pat = MathAbs(sqrt((pow(patternsp - colmat,2.0)).Sum(0))); vector eerep = Dist_pat; np::sort(eerep); int remove = 0; if(targets[i] == 1.0) { vector unq = np::unique(eerep); k = -1; NC1+=1; if(remove!=1) { int Next = 1; while(Next == 1) { k+=1; r = unq[k]; for(ulong j = 0; j<Dist_pat.Size(); j++) { if(Dist_pat[j] == r) f1[j] = 1.0; else f1[j] = 0.0; if(Dist_pat[j]<=r) f2[j] = 1.0; else f2[j] = 0.0; } vector f2t = np::bitwiseAnd(f2,targets); vector tn = np::bitwiseNot(targets); vector f2tn = np::bitwiseAnd(f2,tn); double nocls1clst = f2t.Sum() - 1.0; double nocls2clst = f2tn.Sum(); if(gamma *(nocls1clst/double(n_nt_cls_1-1)) < (nocls2clst/(double(n_nt_cls_2)))) { Next = 0 ; if((k-1) == 0) r = unq[k]; else r = 0.5 * (unq[k-1] + unq[k]); if(r==0.0) r = pow(10.0,-6.0); r = 1.0*r; for(ulong j = 0; j<Dist_pat.Size(); j++) { if(Dist_pat[j]<=r) f2[j] = 1.0; else f2[j] = 0.0; } f2t = np::bitwiseAnd(f2,targets); f2tn = np::bitwiseAnd(f2,tn); nocls1clst = f2t.Sum() - 1.0; nocls2clst = f2tn.Sum(); } } if(Dist_test<r) { patternsp = patterns * np::repeat_vector_as_rows_cols(fs,patterns.Cols(),false); testp = test * fs; dist = MathAbs(sqrt((pow(patternsp - np::repeat_vector_as_rows_cols(testp,patternsp.Cols(),false),2.0)).Sum(0))); min1 = dist; np::sort(min1); min_uniq = np::unique(min1); m = -1; no_nereser = 0; while(no_nereser<int(knn)) { m+=1; double a1 = min_uniq[m]; for(ulong j = 0; j<dist.Size(); j++) NN[j]=(dist[j]<a1)?1.0:0.0; no_nereser = (int)NN.Sum(); } bitNN = np::bitwiseAnd(NN,targets); Not = np::bitwiseNot(targets); NoNNC1[i] = bitNN.Sum(); bitNN = np::bitwiseAnd(NN,Not); NoNNC2[i] = bitNN.Sum(); if(NoNNC1[i]>NoNNC2[i]) S[i] = 1.0; } } } if(targets[i] == 0.0) { vector unq = np::unique(eerep); k=-1; NC2+=1; int Next; if(remove!=1) { Next =1; while(Next==1) { k+=1; r = unq[k]; for(ulong j = 0; j<Dist_pat.Size(); j++) { if(Dist_pat[j] == r) f1[j] = 1.0; else f1[j] = 0.0; if(Dist_pat[j]<=r) f2[j] = 1.0; else f2[j] = 0.0; } vector f2t = np::bitwiseAnd(f2,targets); vector tn = np::bitwiseNot(targets); vector f2tn = np::bitwiseAnd(f2,tn); double nocls1clst = f2t.Sum() ; double nocls2clst = f2tn.Sum() -1.0; if(gamma *(nocls2clst/double(n_nt_cls_2-1)) < (nocls1clst/(double(n_nt_cls_1)))) { Next = 0 ; if((k-1) == 0) r = unq[k]; else r = 0.5 * (unq[k-1] + unq[k]); if(r==0.0) r = pow(10.0,-6.0); r = 1.0*r; for(ulong j = 0; j<Dist_pat.Size(); j++) { if(Dist_pat[j]<=r) f2[j] = 1.0; else f2[j] = 0.0; } f2t = np::bitwiseAnd(f2,targets); f2tn = np::bitwiseAnd(f2,tn); nocls1clst = f2t.Sum(); nocls2clst = f2tn.Sum() -1.0; } } if(Dist_test<r) { patternsp = patterns * np::repeat_vector_as_rows_cols(fs,patterns.Cols(),false); testp = test * fs; dist = MathAbs(sqrt((pow(patternsp - np::repeat_vector_as_rows_cols(testp,patternsp.Cols(),false),2.0)).Sum(0))); min1 = dist; np::sort(min1); min_uniq = np::unique(min1); m = -1; no_nereser = 0; while(no_nereser<int(knn)) { m+=1; double a1 = min_uniq[m]; for(ulong j = 0; j<dist.Size(); j++) NN[j]=(dist[j]<a1)?1.0:0.0; no_nereser = (int)NN.Sum(); } bitNN = np::bitwiseAnd(NN,targets); Not = np::bitwiseNot(targets); NoNNC1[i] = bitNN.Sum(); bitNN = np::bitwiseAnd(NN,Not); NoNNC2[i] = bitNN.Sum(); if(NoNNC2[i]>NoNNC1[i]) S[i] = 1.0; } } } } radious[i] = r; } vector q1 = vector::Zeros(N); vector q2 = vector::Zeros(N); for(int i = 0; i<N; i++) { if(NoNNC1[i] > NoNNC2knn[i]) q1[i] = 1.0; if(NoNNC2[i] > NoNNC1knn[i]) q2[i] = 1.0; } vector ntargs = np::bitwiseNot(targets); vector c1 = np::bitwiseAnd(q1,targets); vector c2 = np::bitwiseAnd(q2,ntargs); double sc1 = c1.Sum()/NC1; double sc2 = c2.Sum()/NC2; if(sc1==0.0 && sc2==0.0) { q1.Fill(0.0); q2.Fill(0.0); for(int i = 0; i<N; i++) { if(NoNNC1knn[i] > NoNNC2knn[i]) q1[i] = 1.0; if(NoNNC2knn[i] > NoNNC1knn[i]) q2[i] = 1.0; if(!targets[i]) ntargs[i] = 1.0; else ntargs[i] = 0.0; } c1 = np::bitwiseAnd(q1,targets); c2 = np::bitwiseAnd(q2,ntargs); sc1 = c1.Sum()/NC1; sc2 = c2.Sum()/NC2; } vector out(2); out[0] = sc1; out[1] = sc2; return out; } public: //+------------------------------------------------------------------+ //| constructor | //+------------------------------------------------------------------+ Clfspy(void) { loaded = false; } //+------------------------------------------------------------------+ //| destructor | //+------------------------------------------------------------------+ ~Clfspy(void) { } //+------------------------------------------------------------------+ //| load a LFSpy trained model from file | //+------------------------------------------------------------------+ bool load(const string file_name, bool FILE_IN_COMMON_DIRECTORY = false) { loaded = false; CFileTxt modelFile; CJAVal js; ResetLastError(); if(modelFile.Open(file_name,FILE_IN_COMMON_DIRECTORY?FILE_READ|FILE_COMMON:FILE_READ,0)==INVALID_HANDLE) { Print(__FUNCTION__," failed to open file ",file_name," .Error - ",::GetLastError()); return false; } else { if(!js.Deserialize(modelFile.ReadString())) { Print("failed to read from ",file_name,".Error -",::GetLastError()); return false; } loaded = fromJSON(js); } return loaded; } //+------------------------------------------------------------------+ //| make a prediction based specific inputs | //+------------------------------------------------------------------+ vector predict(matrix &inputs) { if(!loaded) { Print(__FUNCTION__, " No model available, Load a model first before calling this method "); return vector::Zeros(1); } if(inputs.Cols()!=train_data.Rows()) { Print(__FUNCTION__, " input matrix does np::bitwiseNot match with shape of expected model inputs (columns)"); return vector::Zeros(1); } matrix testdata = inputs.Transpose(); matrix probs = classification(testdata); vector classes = vector::Zeros(probs.Rows()); for(ulong i = 0; i<classes.Size(); i++) if(probs[i][0] > probs[i][1]) classes[i] = 1.0; return classes; } //+------------------------------------------------------------------+ //| get the parameters of the loaded model | //+------------------------------------------------------------------+ LFS_PARAMS getmodelparams(void) { return model_params; } }; //+------------------------------------------------------------------+
There are two primary methods users need to understand in this class:
- The load() method takes a file name as input, which should point to the exported LFS model.
- The predict() method takes a matrix with the requisite number of columns and returns a vector of class labels, corresponding to the number of rows in the input matrix.
Let’s see how all this works in practice. We start with the Python code. The file LFSmodelExportDemo.py prepares in-sample and out-of-sample datasets using randomly generated numbers. The out-of-sample data is saved as a CSV file. An LFS model is trained using the in-sample data, then serialized and saved in JSON format. We test the model on the out-of-sample data and record the results so we can later compare them with the same test done in MetaTrader 5. The Python code is shown next.
# Copyright 2024, MetaQuotes Ltd. # https://www.mql5.com # imports import MetaTrader5 as mt5 import numpy as np import pandas as pd from JsonModel import lfspy2json, LocalFeatureSelection, MQL5_COMMON_FOLDER, MQL5_FILES_FOLDER from os import path from sklearn.metrics import accuracy_score, classification_report #initialize MT5 terminal if not mt5.initialize(): print("MT5 initialization failed ") mt5.shutdown() exit() # stop the script if mt5 not initialized #we want to get the path to the MT5 file sandbox #initialize TerminalInfo instance terminal_info = mt5.terminal_info() #model file name filename = "lfsmodel.json" #build the full path modelfilepath = path.join(terminal_info.data_path,MQL5_FILES_FOLDER,filename) #number of random numbers to generate datalen = 1000 #number of features the dataset will have datavars = 5 #set random number seed rng_seed = 125 rng = np.random.default_rng(rng_seed) #generate the numbers data = rng.uniform(-1.0,1.0,size=datalen) #shape our dataset data = data.reshape([datalen//datavars,datavars]) #set up container for class labels class_labels = np.zeros(shape=data.shape[0],dtype=np.uint8) #set the class labels for i in range(data.shape[0]): class_labels[i] = 1 if (data[i,1] > 0.0 and data[i,2] > 0.0) or (data[i,1] < 0.0 and data[i,2] < 0.0) else 0 #partition our data train_size = 100 xtrain = data[:train_size,:] ytrain = class_labels[:train_size] #load testing data (out of sample) test_data = data[train_size:,:] test_labels = class_labels[train_size:] #here we prepare the out of sample data for export using pandas #the data will be exported in a single csv file colnames = [ f"var_{str(col+1)}" for col in range(test_data.shape[1])] testdata = pd.DataFrame(test_data,columns=colnames) #the last column will be the target labels testdata["c_labels"]=test_labels #display first 5 samples print("Out of sample dataframe head \n", testdata.head()) #display last 5 samples print("Out of sample dataframe tail \n", testdata.tail()) #build the full path of the csv file testdatafilepath=path.join(terminal_info.data_path,MQL5_FILES_FOLDER,"testdata.csv") #try save the file try: testdata.to_csv(testdatafilepath) except Exception as e: print(" Error saving iris test data ") print(e) else: print(" test data successfully saved to csv file ") #initialize the LFS object lfs = LocalFeatureSelection(rr_seed=rng_seed,alpha=8,tau=2,n_beta=20,nrrp=2000) #train the model lfs.fit(xtrain,ytrain) #get the inclusion matrix fstar = lfs.fstar #add up all ones for each row of the inclusion matrix bins = fstar.sum(axis=1) #calculate the percent of times a candidate was selected percents = 100.0 * bins.astype(np.float64)/np.float64(ytrain.shape[0]) index = np.argsort(percents)[::-1] #output the results print("------------------------------> Percent of times selected <------------------------------" ) for i in range(percents.shape[0]): print(f" Variable {colnames[index[i]]}, selected {percents[index[i]]} %") #conduct out of sample test of trained model accuracy = lfs.score(test_data,test_labels) print(f" Out of sample accuracy is {accuracy*100.0} %") #export the model try: lfspy2json(lfs,modelfilepath) except Exception as e: print(" Error saving lfs model ") print(e) else: print("lfs model saved to \n ", modelfilepath)
Next, we shift focus to an MetaTrader 5 script, LFSmodelImportDemo.mq5. Here, we read in the out-of-sample data produced by the Python script and load the trained model. The out-of-sample dataset is then tested, and the results are compared with those obtained from the Python test. The MQL5 code is presented below.
//+------------------------------------------------------------------+ //| LFSmodelImportDemo.mq5 | //| Copyright 2024, MetaQuotes Ltd. | //| https://www.mql5.com | //+------------------------------------------------------------------+ #property copyright "Copyright 2024, MetaQuotes Ltd." #property link "https://www.mql5.com" #property version "1.00" #property script_show_inputs #include<lfspy.mqh> //script inputs input string OutOfSampleDataFile = "testdata.csv"; input bool OutOfSampleDataInCommonFolder = false; input string LFSModelFileName = "lfsmodel.json"; input bool LFSModelInCommonFolder = false; //+------------------------------------------------------------------+ //| Script program start function | //+------------------------------------------------------------------+ void OnStart() { //--- matrix testdata = np::readcsv(OutOfSampleDataFile,OutOfSampleDataInCommonFolder); if(testdata.Rows()<1) { Print(" failed to read csv file "); return; } vector testlabels = testdata.Col(testdata.Cols()-1); testdata = np::sliceMatrixCols(testdata,1,testdata.Cols()-1); Clfspy lfsmodel; if(!lfsmodel.load(LFSModelFileName,LFSModelInCommonFolder)) { Print(" failed to load the iris lfs model "); return; } vector y_pred = lfsmodel.predict(testdata); vector check = MathAbs(testlabels-y_pred); Print("Accuracy is " , (1.0 - (check.Sum()/double(check.Size()))) * 100.0, " %"); } //+------------------------------------------------------------------+
The output from running the Python script LFSmodelExportDemo.py.
Python Out of sample dataframe head Python var_1 var_2 var_3 var_4 var_5 c_labels Python 0 0.337773 -0.210114 -0.706754 0.940513 0.434695 1 Python 1 -0.009701 -0.119561 -0.904122 -0.409922 0.619245 1 Python 2 0.442703 0.295811 0.692888 0.618308 0.682659 1 Python 3 0.694853 0.244405 -0.414633 -0.965176 0.929655 0 Python 4 0.120284 0.247607 -0.477527 -0.993267 0.317743 0 Python Out of sample dataframe tail Python var_1 var_2 var_3 var_4 var_5 c_labels Python 95 0.988951 0.559262 -0.959583 0.353533 -0.570316 0 Python 96 0.088504 0.250962 -0.876172 0.309089 -0.158381 0 Python 97 -0.215093 -0.267556 0.634200 0.644492 0.938260 0 Python 98 0.639926 0.526517 0.561968 0.129514 0.089443 1 Python 99 -0.772519 -0.462499 0.085293 0.423162 0.391327 0 Python test data successfully saved to csv file Python ------------------------------> Percent of times selected <------------------------------ Python Variable var_3, selected 87.0 % Python Variable var_2, selected 81.0 % Python Variable var_4, selected 20.0 % Python Variable var_1, selected 19.0 % Python Variable var_5, selected 18.0 % Python Out of sample accuracy is 92.0 % Python lfs model saved to Python C:\Users\Zwelithini\AppData\Roaming\MetaQuotes\Terminal\FB9A56D617EDDDFE29EE54EBEFFE96C1\MQL5\FILES\lfsmodel.json
Output from running the MQL5 script LFSmodelImportDemo.mq5.
LFSmodelImportDemo (BTCUSD,D1) Accuracy is 92.0 %Comparing the results, we can see that the output from both programs match, indicating that the method of model export works as expected.
Conclusion
Local Feature Selection offers an innovative approach to feature selection, particularly suited for dynamic environments like financial markets. By identifying locally relevant features, LFS overcomes the limitations of traditional methods that rely on a single, global feature set. The algorithm’s adaptability to varying data patterns, its ability to manage non-linear relationships, and its capacity to balance conflicting objectives make it a valuable tool for building machine learning models. While the LFSpy package provides a practical implementation of LFS, there is potential to further optimize its computational efficiency, especially for large-scale datasets. In conclusion, LFS presents a promising approach to classification tasks in domains characterized by complex and evolving data. | File Name | Description |
|---|---|
| Mql5/include/np.mqh | Include file containing generic definitions for various matrix and vector utility functions. |
| Mql5/include/lfspy.mqh | An include file containing definition Clfspy class providing LFS model inference functionality in MetaTrader 5 programs. |
| Mql5/scripts/JsonModel.py | A local Python module containing definition of function enabling export of LFS model in JSON format. |
| Mql5/scripts/LFSdemo.py | A Python script demonstrating how to use the LocalFeatureSelection class for feature selection using random variables |
| Mql5/scripts/LFSmodelExportDemo.py | A Python script demonstrating how to export LFS model for use in MetaTrader 5. |
| Mql5/scripts/LFSmodelImportDemo.mq5 | A MQL5 script showing how to load and use an exported LFS model in a MetaTrader 5 program. |
 Two-sample Kolmogorov-Smirnov test as an indicator of time series non-stationarity
Two-sample Kolmogorov-Smirnov test as an indicator of time series non-stationarity
 Neural Networks Made Easy (Part 87): Time Series Patching
Neural Networks Made Easy (Part 87): Time Series Patching
 MQL5 Wizard Techniques you should know (Part 38): Bollinger Bands
MQL5 Wizard Techniques you should know (Part 38): Bollinger Bands
 How to add Trailing Stop using Parabolic SAR
How to add Trailing Stop using Parabolic SAR
- Free trading apps
- Over 8,000 signals for copying
- Economic news for exploring financial markets
You agree to website policy and terms of use