
Resampling techniques for prediction and classification assessment in MQL5
Introduction
The performance of machine learning models is typically assessed through two distinct phases: training on one dataset and testing with another. However, in situations where collecting multiple datasets may be impractical due to resource constraints or logistical limitations, alternative approaches have to be employed.
One such method involves using resampling techniques to evaluate the performance of prediction or classification models. This approach has been shown to provide reliable results despite its potential drawbacks. In this article, we will explore a novel methodology for assessing model quality that utilizes a single dataset as both training and validation sets. The primary reason for applying these methods is the limited availability of data for testing purposes.
As such, practitioners must employ sophisticated resampling algorithms to produce performance metrics comparable to those generated by more straightforward approaches. These techniques require significant computational resources and may introduce complexity into model development processes. Despite this trade-off, employing resampling-based assessment strategies can be valuable in certain contexts where the benefits outweigh the costs.
Decomposition of errors
To facilitate the exposition of the algorithmic concepts presented in this text, a system of notation is introduced. This notational framework primarily addresses the decomposition of model error into constituent elements. Consider a dataset, denoted as T, derived from a population characterized by an unknown distribution, F. This dataset comprises n observations, each consisting of a predictor variable, x, which may be either scalar or vector-valued, and a predicted variable, y, which is scalar. To emphasize the inherent interdependence of the predictor and predicted variables, the composite term t (for training) is employed to represent the pair (x, y). Thus, the i-th observation in the dataset is represented as t_i = (x_i, y_i). While this notation suggests a numerical prediction framework. Classification problems can be accommodated by interpreting y_i as the class membership of the i-th observation.
The complete dataset is denoted by T. When a model is trained using T as the training set, the resulting trained model is designated as M_T. The application of this trained model, M_T, to a specific value of the predictor variable, x, to generate an estimate of the corresponding predicted variable, yields the model’s prediction, denoted as M_T(x). For the sake of brevity, a generic model prediction may be abbreviated as M. In evaluating the efficacy of a predictive model, it is often necessary to quantify the discrepancy between the model's prediction, M, and the observed value, y, of the predicted variable. This discrepancy is formally defined as an error measure, denoted as Ef[y, M]. A common error measure employed in regression contexts is the mean squared error, which utilizes the squared difference as depicted in the equation below. In classification problems, Ef[y, M] may be defined as a binary function, assigning a value of zero if y is equal to M and one otherwise.
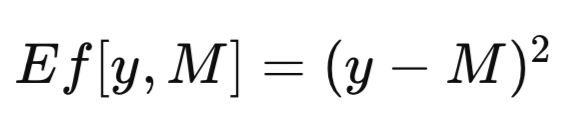
When the trained model is applied to the same dataset used for its training, the mean error, across all training instances is designated as the apparent error. While the conventional approach involves training the model by minimizing this apparent error, the methodology presented herein does not impose such a constraint. The optimization of any arbitrary measure is permissible, followed by the evaluation of the trained model's quality using a distinct criterion. The apparent error, defined below, is solely a function of the model's performance on the training data and the application of the Ef[] function to each prediction; the training methodology is inconsequential.
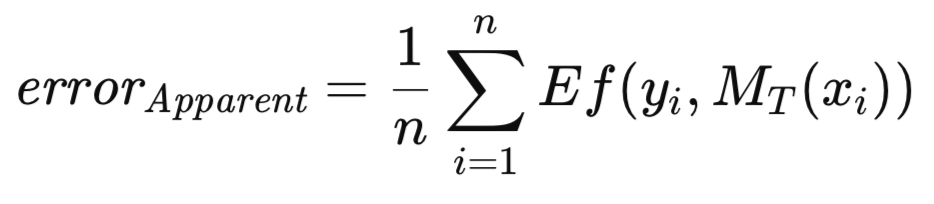
It is well-established that the apparent error exhibits an optimistic bias, a phenomenon sometimes referred to as training bias, due to the model's evaluation on the same data used for its training. To mitigate this bias, when sufficient data is available, the trained model is evaluated on an independent dataset. This procedure facilitates the estimation of the trained model's expected error when applied to the general population. This expected error, conditioned on the model having been trained on the training set, is termed the prediction error. The emphasis here is on the evaluation of a specific trained model's anticipated future performance. The prediction error is formally expressed in the following equation.
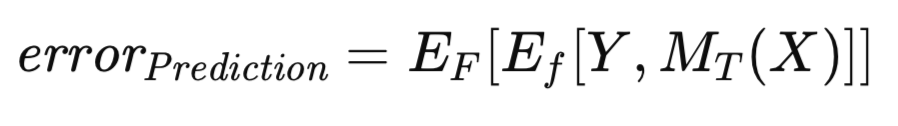
A related, yet conceptually distinct, measure of error warrants consideration. The prediction error, as previously defined, is conditional upon a specific realized model trained on a particular dataset. However, it is possible to extend the expectation to encompass the ensemble of all possible training sets. Specifically, given that the training set comprises n observations sampled from an unknown distribution, one can assess the prediction error associated with the model trained on this particular training dataset, potentially approximating it via an independent validation set. Now, consider the scenario where a new training set is drawn, and the training procedure is reiterated. Inevitably, a slightly divergent prediction error would be obtained. Consequently, it becomes necessary to formulate a measure that captures the expected prediction error across the entire spectrum of potential training sets. This measure, termed the population error, is formally defined below.
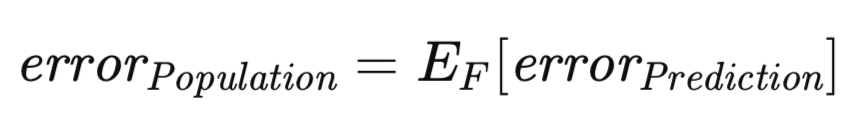
It is a well-established principle that, for any given training set, the apparent error will, with high probability, underestimate the prediction error. This discrepancy arises from the model's tendency to overfit the specific characteristics of the training data, thereby compromising its generalizability to the broader population. The magnitude of the difference between the prediction error and the apparent error is defined as the excess error.

Of particular significance in this context is the expected excess error. Recognizing that both the prediction error and the apparent error are contingent upon a specific training set, it is necessary to consider the expected value of the excess error across the ensemble of all possible training sets, analogous to the derivation of the population error. This is formally represented in the equation below.
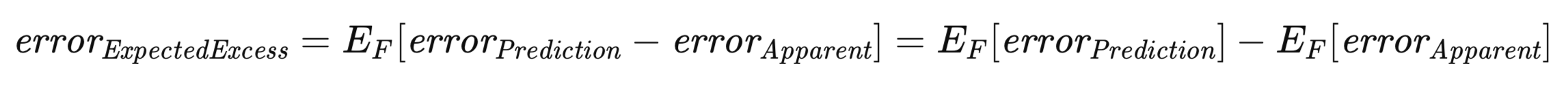
The equation above underscores the duality in conceptualizing the expected excess error. It can be viewed as the expectation, across all possible training sets, of the difference between the prediction error and the apparent error for each individual training set. This interpretation aligns with an intuitive understanding of the concept. Alternatively, it can be expressed as the difference between the population error and the expected apparent error.
Cross-Validation: Methodology and limitations
Cross-validation is a well-established technique within the research community. Among the algorithms discussed in this text, cross-validation is notable for its conceptual simplicity and ease of implementation, often exhibiting computational efficiency. It possesses the advantageous characteristic of yielding a nearly unbiased estimate of a model's expected future performance.
Furthermore, its broad applicability extends to a wide range of model training algorithms, a feature not universally shared by other methods. However, cross-validation is subject to a significant limitation: its inherent variance is frequently substantial, potentially to an unacceptable degree. This implies a heightened sensitivity to the stochastic variations introduced by the random sampling of the original dataset.
Specifically, an experimenter who collects a sample, trains a model, and subsequently employs cross-validation to assess the model's expected future performance may observe a markedly different result upon repeating the procedure with an independent sample. While the near-unbiased nature of the estimate is a desirable property, it is often overshadowed by the magnitude of the variance. Critically, this variability is frequently underestimated by practitioners.
Despite the existence of superior methodologies for analogous tasks, cross-validation remains widely employed, often due to its simplicity and its applicability in scenarios where alternative approaches are infeasible. Nonetheless, given its prevalence and occasional necessity, a detailed exposition of the cross-validation procedure is warranted.
The fundamental principle underlying cross-validation is conceptually straightforward. It involves partitioning the dataset into two distinct subsets: a training set, used for model parameter estimation, and a validation set, employed for independent model evaluation, mirroring the procedure adopted when data abundance is not a constraint.
However, in contrast to allocating a substantial portion of the data to the validation set for robust performance estimation, cross-validation utilizes a minimal validation set, allocating the majority of observations to the training set. Specifically, it is common practice to employ a single observation as the validation set.
Following the training and evaluation of the model with this partition, the validation set is reintegrated into the dataset, and a different observation is designated as the validation set. This process is iterated until each observation has served as a validation point once. The mean test error across all iterations constitutes the cross-validation error estimate.
The most prevalent variant of cross-validation involves the sequential exclusion of single observations. The adaptation of these algorithms for the exclusion of multiple observations should be easy enough. Given that M_T represents the model trained on the complete dataset, T, let M_T(i) denote the model trained on the dataset T excluding the i-th observation. The cross-validation estimate of the model's expected future error is then formally expressed as follows.
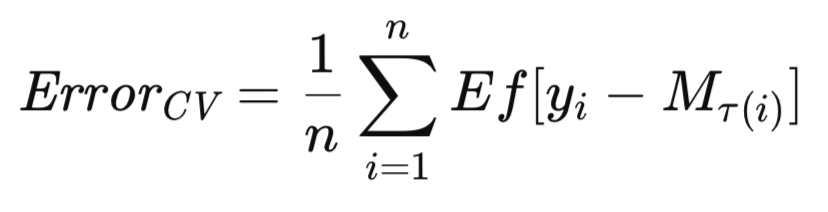
This section presents the cross-validation algorithm, accompanied by explanatory notes. Comparative performance evaluations are provided towards the end of this article. All the algorithms used to estimate expected errors are presented in the header file, error_variance_estimation.mqh. The cross-validation algorithm is implemented as the method cross_validation(). This routine and those used to implement other error estimation algorithms have a similar structure and function parameters. These parameters are:
- The first two required arguments to the method are matrices of the training dataset. The first matrix holds the predictors and the second, the corresponding targets.
- The third argument to the routine is an instance of a model, that implements the IModel interface. This represents the prediction model being evaluated.
- The last argument passed to the cross_validation() holds the final error measure obtained from the computation. If an error occurs the method will return the boolean value, false.
//+------------------------------------------------------------------+ //| estimate error variance using cross validation testing | //+------------------------------------------------------------------+ bool CErrorVar::cross_validation(matrix &predictors, matrix &targets, IModel & model,double &out_err) { out_err = 0.0; vector test; double test_target; matrix preds,targs; for(ulong i = 0; i<predictors.Rows(); i++) { test = predictors.Row(i); test_target = targets[i][0]; predictors.SwapRows(i,(predictors.Rows()-1)); targets.SwapRows(i,(targets.Rows()-1)); preds = np::sliceMatrixRows(predictors,0,long(predictors.Rows()-1)); targs = np::sliceMatrixRows(targets,0,long(targets.Rows()-1)); if(!model.train(preds,targs)) { Print(__FUNCTION__," failed to train model "); return false; } out_err += error_fun(test_target,model.forecast(test)); predictors.SwapRows(i,(predictors.Rows()-1)); targets.SwapRows(i,(targets.Rows()-1)); } out_err/=double(predictors.Rows()); return true; }
Invocation of cross_validation() triggers the commencement of a loop that traverses the training dataset. The last position in the training dataset is always used as the test spot. The loop swaps other samples into this position in the predictor and target matrices, one at a time. Once the data is partitioned, the model is trained and then tested on the single sample that was excluded from training. Its error is cumulated and eventually returned as the result of going through the whole loop.
Bootstrap estimation of population error
This section delineates a basic bootstrap algorithm for the estimation of population error. It is acknowledged that this algorithm is not generally recommended for practical applications, as the E0 and E632 algorithms, detailed in subsequent sections, typically offer superior performance. Nevertheless, the direct bootstrap method presented here serves as the foundational principle upon which more sophisticated algorithms are constructed. Thus, a comprehensive understanding of its operational mechanics is indispensable for comprehending its successors.
In the present scenario, we posit a population, F, from which the observations comprising our training set are randomly sampled. The model is trained and evaluated on T, yielding the apparent error. This error is inherently optimistic, and the population error is expected to exceed the apparent error by the excess error. However, in the absence of an independent validation set, neither the excess error nor the population error, the quantities of primary interest, are directly observable. We are limited to the optimistic apparent error. Nevertheless, the bootstrap methodology can be employed to estimate the excess error, which can then be added to the apparent error to provide a rough estimate of the population error.
The utilization of the bootstrap for excess error estimation mirrors the procedure for estimating parameter bias. An empirical distribution function, is substituted for the unknown population distribution and numerous bootstrap samples are drawn. For each bootstrap sample, the model is trained. The model's error on the bootstrap sample represents the apparent error for that sample. The model's error on the complete dataset represents the prediction error, as the empirical distribution effectively functions as the entire population. The difference between these two errors yields the excess error for that bootstrap sample. The average of this excess error across numerous bootstrap replications provides an estimate of the expected excess error.
To enhance the rigor of the aforementioned algorithm, a series of definitions and equations are introduced. Let B denote a training set generated through bootstrap sampling from the original dataset. Specifically, B is constructed by randomly selecting n observations from T with replacement. The model is subsequently trained using B. The prediction error of this model is determined by averaging its error across all observations in the population from which the bootstrap sample was drawn, which, in this context, is the original dataset. This is formally represented in the equation below.
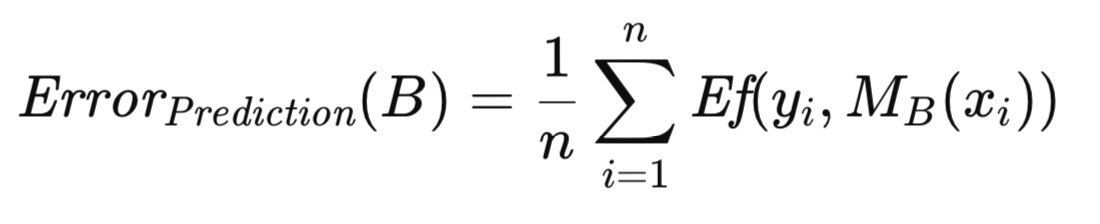
Let k_i represent the frequency with which the i-th observation from T appears in B. The apparent error associated with B is computed by averaging the model's error across the observations comprising B, as shown in the equation below.
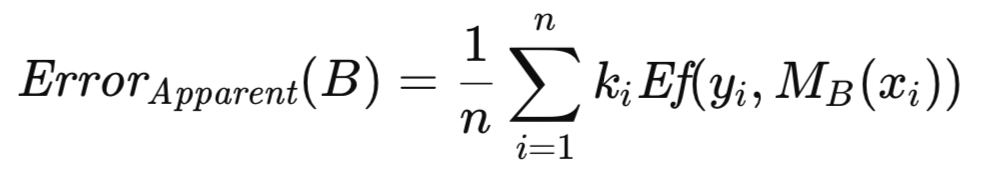
The excess error associated with B is defined as the difference between its prediction error and its apparent error. Rather than computing them separately and then subtracting, which would be computationally redundant due to the presence of shared error terms, a more efficient formulation is obtained by factoring out the common quantity. This results in an expression that must be evaluated for each of a substantial number (on the order of hundreds to thousands) of bootstrap replications. The average excess error across these replications provides an estimate of the expected excess error, which is then added to the apparent error of the full sample to yield an approximation of the population error.
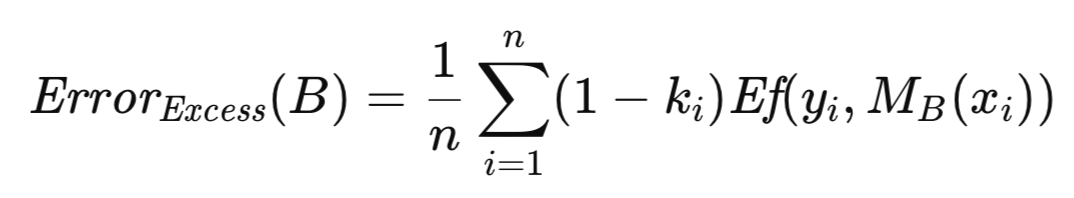
The bootstrap technique for error estimation is implemented as the method boot_strap(). In addition to the parameters listed for the cross-validation implementation, this method includes an additional parameter, specifying the number of bootstrap replications. The routine starts by initializing an instance of a random number generator courtesy of the MQL5 implementation of the Alglib library. The random number generator is used to select a random row index as a bootstrap sample that is placed in the matrix variables 'preds' and 'targs' from the original training set. The bootstrap sample set is used to train the model and subsequently test it, to accumulate the excess error.
//+------------------------------------------------------------------+ //| estimate error variance using ordinary bootstrap | //+------------------------------------------------------------------+ bool CErrorVar::boot_strap(ulong nboot,matrix &predictors, matrix &targets, IModel & model,double &out_err) { double err,apparent,excess; excess = 0.0; ulong nsize = predictors.Rows(); ulong count[]; ulong k; ArrayResize(count,int(nsize)); vector predicted(nsize); CHighQualityRandStateShell rstate; CHighQualityRand::HQRndRandomize(rstate.GetInnerObj()); matrix preds = predictors; matrix targs = targets; for(ulong boot = 0; boot<nboot; boot++) { ArrayInitialize(count,0); //--- for(ulong i=0; i<nsize; i++) { k=(int)(CAlglib::HQRndUniformR(rstate)*nsize); //--- if(k>=nsize) k=nsize-1; //--- preds.Row(predictors.Row(k),i); targs.Row(targets.Row(k),i); ++count[k]; } if(!model.train(preds,targs)) { Print(__FUNCTION__," failed to train model ", boot); return false; } for(ulong i=0; i<nsize; i++) { predicted[i] = model.forecast(predictors.Row(i)); err = error_fun(targets[i][0],predicted[i]); excess+=(1.0 - double(count[i]))*err; } } excess/=double(nsize*nboot);
Once the bootstrap operation is completed, the original training dataset is used to calculate the apparent error. The final error estimate becomes the combined value of the excess and apparent errors.
if(!model.train(predictors,targets)) { Print(__FUNCTION__," failed to train model "); return false; } apparent = 0.0; for(ulong i=0; i<nsize; i++) { predicted[i] = model.forecast(predictors.Row(i)); err = error_fun(targets[i][0],predicted[i]); apparent+=err; } apparent/=double(nsize); out_err = apparent+excess; return true; }
Efron's E0 estimator for population error
A notable challenge arising from the duplication of training cases within bootstrap samples is the potential for rendering certain model classes inoperative. Specifically, probabilistic and generalized regression neural networks are particularly susceptible. Unless the smoothing constant is sufficiently large, a test case identical to a training case will yield a near-perfect prediction, exacerbating the issue of optimistic bias. To mitigate this problem, Bradley Efron proposed a straightforward solution: the prevention of duplication artifacts. This involves generating bootstrap training sets through standard resampling procedures. However, for each bootstrap replication, the model is evaluated solely on those original observations that are absent from the training set. The mean error across these excluded observations is then employed as an estimate of the population error. This methodology is designated as the E0 estimator of population error.
Academic literature presents two distinct approaches for computing E0. The original method, involves summing all errors and dividing by the total number of evaluated cases. This approach is adopted herein. Subsequent theoretical investigations into the properties of E0, propose an alternative algorithm that decomposes the averaging process into two stages. First, for each original observation, the errors from all bootstrap replications lacking that observation are summed and divided by the number of such replications, yielding a mean error for that observation. Subsequently, these mean errors are summed across all observations and divided by the total number of observations to obtain a grand mean. While this latter method exhibits increased computational complexity, it is asymptotically equivalent to the former, and empirical evaluations demonstrate negligible performance differences. Consequently, the original method is selected for its simplicity.
To provide a better description of the algorithm, additional notation is introduced. As before, T denotes the original dataset, and B represents a bootstrap sample. Let C denote the set of observations in T that are not included in B. Let count(C) represent the cardinality (number of observations) of C. Then, Efron's original E0 estimate of population error is formally expressed below.
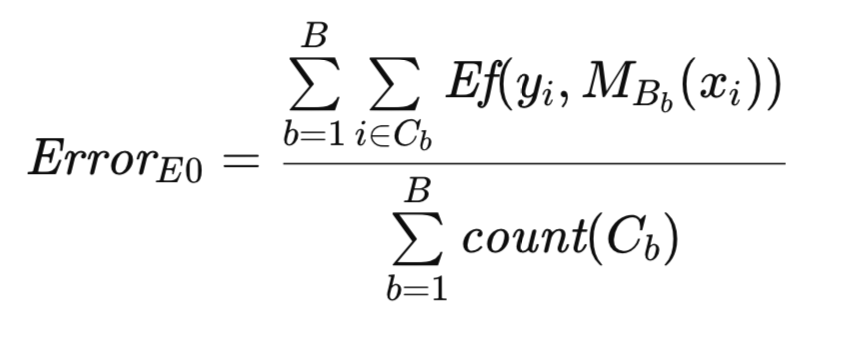
The algorithmic implementation of the E0 estimator bears resemblance to the previously described bootstrap routine. The generation and utilization of bootstrap samples for model training are consistent across both methods. However, in this context, the count array serves as a binary indicator, representing the presence or absence of an observation, rather than as a frequency counter as in the preceding routine. The algorithm is implemented as the method efrons_0().
//+------------------------------------------------------------------+ //| estimate error variance using efron's E0 bootstrap | //+------------------------------------------------------------------+ bool CErrorVar::efrons_0(ulong nboot,matrix &predictors, matrix &targets, IModel & model,double &out_err) { out_err = 0.0; ulong tot = 0; ulong nsize = predictors.Rows(); ulong count[]; ulong k; ArrayResize(count,int(nsize)); vector predicted(nsize); CHighQualityRandStateShell rstate; CHighQualityRand::HQRndRandomize(rstate.GetInnerObj()); matrix preds = predictors; matrix targs = targets; for(ulong boot = 0; boot<nboot; boot++) { ArrayInitialize(count,0); //--- for(ulong i=0; i<nsize; i++) { k=(int)(CAlglib::HQRndUniformR(rstate)*nsize); //--- if(k>=nsize) k=nsize-1; //--- preds.Row(predictors.Row(k),i); targs.Row(targets.Row(k),i); ++count[k]; } if(!model.train(preds,targs)) { Print(__FUNCTION__," failed to train model ", boot); continue;//return false; } for(ulong i=0; i<nsize; i++) { if(count[i]) continue; predicted[i] = model.forecast(predictors.Row(i)); out_err+= error_fun(targets[i][0],predicted[i]); ++tot; } } if(tot) out_err/=double(tot); else { Print(__FUNCTION__, " zero denominator "); return false; } return true; }
Efron's E632 estimator for population error
The E0 estimator, as previously described, exhibits desirable properties and is generally recommended for practical application. Its avoidance of evaluating observations present in the training set ensures its compatibility with diverse model classes. Furthermore, its variance is reasonably constrained, reflecting the current state of methodological development.
However, the E0 estimator is characterized by a moderate conservative bias, tending to overestimate the true population error. It is important to note that, provided this bias is not excessive, it is generally considered less problematic than underestimation. Underestimation, as observed with the ordinary bootstrap, is more detrimental due to its propensity to foster unwarranted optimism. The utilization of the E0 method typically provides assurance that the actual population error is more likely to be lower than the computed value. This inherent conservatism, while generally advantageous, may lead to the erroneous rejection of a model due to undue pessimism. The E632 estimator is designed to address this issue by mitigating the conservative bias inherent in E0.
Consider the inherent limitations of employing the apparent error (the error obtained from evaluating the training set) as an estimate of population error. The evaluation procedure is inherently biased, as only observations utilized in training are subject to evaluation. This subset is not representative of the entire population, exhibiting excessive similarity to the training set, resulting in optimistic bias.
Conversely, the E0 estimator exhibits the opposite bias. By deliberately excluding training observations from evaluation, the test set becomes unrepresentative of the population, exhibiting excessive dissimilarity to the training set. In real-world scenarios, observations identical or nearly identical to training observations will inevitably arise. E0's exclusion of these observations leads to pessimistic bias.
The E632 algorithm seeks to establish a compromise between these two extremes. A more equitable approach might involve evaluating the model by sampling both within and outside the training set, with probabilities reflecting their real-world occurrence. Alternatively, adjustments can be made to account for sampling discrepancies. As the sample size increases, the probability of any given observation appearing in a bootstrap sample converges to 1 − 1/e ≈ 0.632. Efron's heuristic proposes estimating the population error as a weighted sum of E0 and the apparent error, with weights determined by these sampling probabilities. His estimator, designated E632, is formally represented in the equation below.
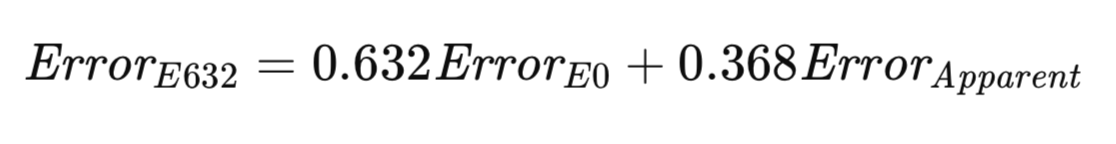
The E632 algorithm implementation is provided as the method efrons_632(), which is shown next.
//+------------------------------------------------------------------+ //| estimate error variance using efron's E632 bootstrap | //+------------------------------------------------------------------+ bool CErrorVar::efrons_632(ulong nboot,matrix &predictors, matrix &targets, IModel & model,double &out_err) { double apparent; if(!efrons_0(nboot,predictors,targets,model,out_err)) return false; if(!model.train(predictors,targets)) { Print(__FUNCTION__," failed to train model "); return false; } apparent = 0.0; vector predicted(predictors.Rows()); for(ulong i=0; i<predictors.Rows(); i++) { predicted[i] = model.forecast(predictors.Row(i)); apparent+= error_fun(targets[i][0],predicted[i]); } apparent/=double(predictors.Rows()); out_err = 0.632*out_err + 0.368*apparent; return true; }
All the algorithms discussed in this text so far are implemented as members of the CErrorVar class. This class also defines the error_fun() method used to compute the error between a predicted value, provided as a second parameter and the corresponding target value, supplied as the method's first parameter.
//+------------------------------------------------------------------+ //| class for estimating error variance | //+------------------------------------------------------------------+ class CErrorVar { public: CErrorVar(void); ~CErrorVar(void); virtual double error_fun(const double truevalue,const double predictedvalue); virtual bool cross_validation(matrix &predictors, matrix &targets, IModel & model,double &out_err); virtual bool boot_strap(ulong nboot,matrix &predictors, matrix &targets, IModel & model,double &out_err); virtual bool efrons_0(ulong nboot,matrix &predictors, matrix &targets, IModel & model,double &out_err); virtual bool efrons_632(ulong nboot,matrix &predictors, matrix &targets, IModel & model,double &out_err); }; //+------------------------------------------------------------------+ //| constructor | //+------------------------------------------------------------------+ CErrorVar::CErrorVar(void) { } //+------------------------------------------------------------------+ //| destructor | //+------------------------------------------------------------------+ CErrorVar::~CErrorVar(void) { } //+------------------------------------------------------------------+ //| calculate the error | //+------------------------------------------------------------------+ double CErrorVar::error_fun(const double truevalue,const double predictedvalue) { return pow(truevalue-predictedvalue,2.0); }
The next section provides a demonstration of these algorithms being used to estimate the error of a trained model using only the training dataset.
Comparative analysis of prediction error estimators
This text has presented a suite of methodologies for estimating the population error of a predictive model. The following key considerations should be noted:
- Cross-Validation: This technique is characterized by its ease of implementation and computational efficiency. It exhibits broad applicability across diverse model classes and provides a nearly unbiased estimate. However, it is susceptible to high variance, particularly in scenarios involving unstable training procedures. Consequently, cross-validation is generally not recommended as a primary method unless alternative approaches are unavailable. While effective, it is not considered optimal.
- Straight Bootstrap: This method is generally considered the least desirable option. It is incompatible with models that cannot accommodate duplicate training observations or those compromised by the presence of test observations within the training set. Furthermore, it exhibits a significant bias towards underestimating the true population error. Therefore, it lacks compelling advantages.
- E0 Estimator: In scenarios where the training process allows for duplicate observations, the E0 estimator is generally the preferred choice. Its applicability extends to a wide range of models, as it avoids evaluating training observations. It demonstrates robustness in the presence of unstable learning conditions, such as those involving models with stochastic training procedures. Empirically, its variance approximates that of cross-validation in stable learning environments, and it exhibits significantly reduced variance in unstable environments. Furthermore, it is computationally efficient, given that the apparent error, required for the E632 estimator, can be computed concurrently with the E0 estimate. However, it is important to acknowledge that practitioners may prioritize the conservative bias of E0 over the potentially higher variance associated with the less biased E632 estimator.
To empirically evaluate these estimators, a simulation study was conducted using artificial data generated according to the model specified as, y = x_1 - x_2 + error. In this model, the predictor variables, x_1 and x_2, follow a standard normal distribution, and the error term, is normally distributed with a mean of zero and a variance specified by the user. An ordinary linear model was fitted to the dataset, and the population mean squared error was estimated using the algorithms presented in this article. The fitted model was also evaluated on independent test data to determine its true error. This process was repeated for a user-specified number of trials, and the average and standard deviation of the error estimates were calculated. This is implemented in the script, ErrorVarianceEstimation_NumericalPredictionDemo.mq5.
//+------------------------------------------------------------------+ //| ErrorVarianceEstimation_NumericalPredictionDemo.mq5 | //| Copyright 2024, MetaQuotes Ltd. | //| https://www.mql5.com | //+------------------------------------------------------------------+ #property copyright "Copyright 2024, MetaQuotes Ltd." #property link "https://www.mql5.com" #property version "1.00" #property script_show_inputs #include<error_variance_estimation.mqh> #include<OLS.mqh> //--- input parameters input ulong NumSamples=15; input ulong NumBootStraps = 1000; input ulong NumReplications = 100; input double Variance = 1.0; //--- //+------------------------------------------------------------------+ //| normal(rngstate) | //+------------------------------------------------------------------+ double normal(CHighQualityRandStateShell &state) { return CAlglib::HQRndNormal(state); } //+------------------------------------------------------------------+ //| unifrand(rngstate) | //+------------------------------------------------------------------+ double unifrand(CHighQualityRandStateShell &state) { return CAlglib::HQRndUniformR(state); } //+------------------------------------------------------------------+ //| ordinary least squares class | //+------------------------------------------------------------------+ class COrdReg:public IModel { private: OLS* m_ols; public: COrdReg(void) { m_ols = new OLS(); } ~COrdReg(void) { if(CheckPointer(m_ols) == POINTER_DYNAMIC) delete m_ols; } bool train(matrix &predictors,matrix& targets) { return m_ols.Fit(targets.Col(0),predictors); } double forecast(vector &predictors) { return m_ols.Predict(predictors); } }; //--- ulong nreplications, itry, nsamps, nboots, divisor, ndone; vector computed_err_cv, computed_err_boot, predictions; vector computed_err_E0, computed_err_E632 ; double temperr,sum_observed_error, mean_computed_err, var_computed_err,dfactor,dif; matrix xdata, testdata,trainpreds,traintargs,testpreds,testtargs; //+------------------------------------------------------------------+ //| Script program start function | //+------------------------------------------------------------------+ void OnStart() { CHighQualityRandStateShell rngstate; CHighQualityRand::HQRndRandomize(rngstate.GetInnerObj()); //--- nboots = NumBootStraps; nsamps = NumSamples ; nreplications = NumReplications ; dfactor = Variance ; if((nsamps <= 3) || (nreplications <= 0) || (dfactor < 0.0) || nboots<=0) { Alert(" Invalid inputs "); return; } double std = sqrt(dfactor) ; divisor = 1000000 / (nsamps * nboots) ; // This is for progress reports only if(divisor < 2) divisor = 2 ; xdata = matrix::Zeros(nsamps,3); sum_observed_error = mean_computed_err = var_computed_err = 0.0; computed_err_cv = vector::Zeros(nreplications); computed_err_E0 = vector::Zeros(nreplications); computed_err_E632 = vector::Zeros(nreplications); computed_err_boot = vector::Zeros(nreplications); testdata = matrix::Zeros(nsamps*10,3); predictions = vector::Zeros(nsamps*10); CErrorVar errorvar; COrdReg regmodel; for(ulong irep = 0; irep<nreplications; irep++) { ndone = irep + 1 ; for(ulong i =0; i<nsamps; i++) { xdata[i][0] = normal(rngstate); xdata[i][1] = normal(rngstate); xdata[i][2] = xdata[i][0] - xdata[i][1] + std * normal(rngstate); } for(ulong j =0; j<testdata.Rows(); j++) { testdata[j][0] = normal(rngstate); testdata[j][1] = normal(rngstate); testdata[j][2] = testdata[j][0] - testdata[j][1] + std *normal(rngstate); } trainpreds = np::sliceMatrixCols(xdata,0,2); traintargs = np::sliceMatrixCols(xdata,2); if(!regmodel.train(trainpreds,traintargs)) { Print(" fitting first model failed "); return; } testpreds=np::sliceMatrixCols(testdata,0,2); testtargs=np::sliceMatrixCols(testdata,2); temperr = 0.0; for(ulong i = 0;i<testpreds.Rows(); i++) { predictions[i] = regmodel.forecast(testpreds.Row(i)); temperr += errorvar.error_fun(testtargs[i][0],predictions[i]); } sum_observed_error += temperr/double(10*nsamps); if(!errorvar.cross_validation(trainpreds,traintargs,regmodel,computed_err_cv[irep]) || !errorvar.boot_strap(nboots,trainpreds,traintargs,regmodel,computed_err_boot[irep]) || !errorvar.efrons_0(nboots,trainpreds,traintargs,regmodel,computed_err_E0[irep]) || !errorvar.efrons_632(nboots,trainpreds,traintargs,regmodel,computed_err_E632[irep]) ) { Print(" error variance calculation failed "); return; } //--- } //--- PrintFormat("Number of Iterations %d Observed error = %.5lf",ndone, sum_observed_error / double(ndone)) ; //--- PrintFormat("CV: computed error mean=%10.5lf std=%10.5lf",computed_err_cv.Mean(), computed_err_cv.Std()) ; //--- PrintFormat("BOOT: computed error mean=%10.5lf std=%10.5lf",computed_err_boot.Mean(), computed_err_boot.Std()) ; //--- PrintFormat("E0: computed error mean=%10.5lf std=%10.5lf",computed_err_E0.Mean(), computed_err_E0.Std()) ; //--- PrintFormat("E632: computed error mean=%10.5lf std=%10.5lf",computed_err_E632.Mean(), computed_err_E632.Std()) ; } //+------------------------------------------------------------------+
Using variance of 1.0, a sample size of 15 observations, and 1,000 bootstrap iterations. The results were as follows:
MJ 0 12:40:47.575 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) Number of Iterations 100 Observed error = 1.18380 RF 0 12:40:47.575 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) CV: computed error mean= 1.18825 std= 0.53117 PK 0 12:40:47.575 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) BOOT: computed error mean= 1.12521 std= 0.48780 IR 0 12:40:47.575 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) E0: computed error mean= 1.38168 std= 0.63579 NO 0 12:40:47.575 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) E632: computed error mean= 1.18647 std= 0.52380
As anticipated, the standard bootstrap method exhibited an underestimation of the true error. Conversely, the E0 estimator significantly overestimated the error. While this overestimation may be perceived as a limitation, it is important to note that the E0 estimator also exhibited the highest standard deviation. This degree of overestimation is primarily attributed to the tiny sample size. However, the decision regarding its acceptability remains subjective. To further evaluate the performance of the error estimators, the simulation study was replicated with an increased sample size of 100 observations, a more representative scenario for many practical applications. Here are the results:
KG 0 12:43:23.483 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) CV: computed error mean= 1.01810 std= 0.24132 LH 0 12:43:23.483 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) BOOT: computed error mean= 1.01672 std= 0.14194 PS 0 12:43:23.483 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) E0: computed error mean= 1.01989 std= 0.14441 IP 0 12:43:23.483 ErrorVarianceEstimation_NumericalPredictionDemo (Gold RSI Trend Up Index,H1) E632: computed error mean= 1.01855 std= 0.14099
The results of this experiment demonstrate satisfactory performance across all four methodologies. Notably, the E0 estimator exhibited a slight overestimation of the error. The remaining three algorithms exhibited negligible underestimation. While the E0 estimator maintained the highest standard deviation, the difference was minimal. In scenarios characterized by high overall estimation quality, the minor overestimation of error produced by the E0 estimator is likely to be preferred over the trivial underestimation observed in the other methods, particularly given the implications of underestimation for model selection and evaluation.
The preceding example illustrated certain distinctions among the various methods for estimating population error. However, it may have inadvertently conveyed the impression that cross-validation is consistently comparable to other methods in terms of effectiveness. This perception is attributable to the employment of a smooth error function, specifically the mean-squared error of a simple model, which fosters a stable learning environment.
Classification tasks, in contrast, are generally characterized by inherent instability. Minor perturbations in the data can precipitate abrupt and substantial fluctuations in the error rate. This section presents an example that explores this phenomenon. The algorithms employed for estimating population error remain consistent with those previously discussed. The principal modifications pertain to the definition of the prediction error function and the structure of the main program used for comparative evaluation.
The test script, ErrorVarianceEstimation_ClassificationDemo.mq5, generates bivariate data exhibiting moderate positive correlation.
//+------------------------------------------------------------------+ //| ErrorVarianceEstimation_ClassificationDemo.mq5 | //| Copyright 2024, MetaQuotes Ltd. | //| https://www.mql5.com | //+------------------------------------------------------------------+ #property copyright "Copyright 2024, MetaQuotes Ltd." #property link "https://www.mql5.com" #property version "1.00" #property script_show_inputs #include<error_variance_estimation.mqh> #include<OLS.mqh> //--- input parameters input ulong NumSamples=15; input ulong NumBootStraps = 1000; input ulong NumReplications = 100; input double PredictionDifficultyLevel = 0.0; //--- //+------------------------------------------------------------------+ //| normal(rngstate) | //+------------------------------------------------------------------+ double normal(CHighQualityRandStateShell &state) { return CAlglib::HQRndNormal(state); } //+------------------------------------------------------------------+ //| unifrand(rngstate) | //+------------------------------------------------------------------+ double unifrand(CHighQualityRandStateShell &state) { return CAlglib::HQRndUniformR(state); } //+------------------------------------------------------------------+ //| ordinary least squares class | //+------------------------------------------------------------------+ class COrdReg:public IModel { private: OLS* m_ols; public: COrdReg(void) { m_ols = new OLS(); } ~COrdReg(void) { if(CheckPointer(m_ols) == POINTER_DYNAMIC) delete m_ols; } bool train(matrix &predictors,matrix& targets) { return m_ols.Fit(targets.Col(0),predictors); } double forecast(vector &predictors) { return m_ols.Predict(predictors); } }; //+------------------------------------------------------------------+ //| error variance for classification models | //+------------------------------------------------------------------+ class CErrorVarC:public CErrorVar { public: CErrorVarC(void) { } ~CErrorVarC(void) { } virtual double error_fun(const double truevalue,const double predictedvalue) { if(truevalue*predictedvalue>0.0) return 0.0; else return 1.0; } }; //--- ulong nreplications, itry, nsamps, nboots, divisor, ndone; vector computed_err_cv, computed_err_boot, predictions; vector computed_err_E0, computed_err_E632 ; double temperr,sum_observed_error, mean_computed_err, var_computed_err,dfactor,dif; matrix xdata, testdata,trainpreds,traintargs,testpreds,testtargs; //+------------------------------------------------------------------+ //| Script program start function | //+------------------------------------------------------------------+ void OnStart() { CHighQualityRandStateShell rngstate; CHighQualityRand::HQRndRandomize(rngstate.GetInnerObj()); //--- nboots = NumBootStraps; nsamps = NumSamples ; nreplications = NumReplications ; dfactor = PredictionDifficultyLevel ; if((nsamps <= 3) || (nreplications <= 0) || (dfactor < 0.0) || nboots<=0) { Alert(" Invalid inputs "); return; } double std = sqrt(dfactor) ; divisor = 1000000 / (nsamps * nboots) ; // This is for progress reports only if(divisor < 2) divisor = 2 ; xdata = matrix::Zeros(nsamps,3); sum_observed_error = mean_computed_err = var_computed_err = 0.0; computed_err_cv = vector::Zeros(nreplications); computed_err_E0 = vector::Zeros(nreplications); computed_err_E632 = vector::Zeros(nreplications); computed_err_boot = vector::Zeros(nreplications); testdata = matrix::Zeros(nsamps*10,3); predictions = vector::Zeros(nsamps*10); CErrorVarC errorvar; COrdReg olsmodel; for(ulong irep = 0; irep<nreplications; irep++) { ndone = irep + 1 ; for(ulong i =0; i<nsamps; i++) { xdata[i][0] = normal(rngstate); xdata[i][1] = 0.7071 * xdata[i][0] + 0.7071 * normal(rngstate); if(CAlglib::HQRndUniformR(rngstate)>0.5) { xdata[i][0] -=dfactor; xdata[i][1] +=dfactor; xdata[i][2] = 1.0; } else { xdata[i][0] +=dfactor; xdata[i][1] -=dfactor; xdata[i][2] = -1.0; } } for(ulong j =0; j<testdata.Rows(); j++) { testdata[j][0] = normal(rngstate); testdata[j][1] = 0.7071 * testdata[j][0] + 0.7071 * normal(rngstate); if(CAlglib::HQRndUniformR(rngstate)>0.5) { testdata[j][0] -=dfactor; testdata[j][1] +=dfactor; testdata[j][2] = 1.0; } else { testdata[j][0] +=dfactor; testdata[j][1] -=dfactor; testdata[j][2] = -1.0; } } trainpreds = np::sliceMatrixCols(xdata,0,2); traintargs = np::sliceMatrixCols(xdata,2); if(!olsmodel.train(trainpreds,traintargs)) { Print(" fitting first model failed "); return; } testpreds=np::sliceMatrixCols(testdata,0,2); testtargs=np::sliceMatrixCols(testdata,2); temperr = 0.0; for(ulong i = 0;i<testpreds.Rows(); i++) { predictions[i] = olsmodel.forecast(testpreds.Row(i)); temperr += errorvar.error_fun(testtargs[i][0],predictions[i]); } sum_observed_error += temperr/double(10*nsamps); if(!errorvar.cross_validation(trainpreds,traintargs,olsmodel,computed_err_cv[irep]) || !errorvar.boot_strap(nboots,trainpreds,traintargs,olsmodel,computed_err_boot[irep]) || !errorvar.efrons_0(nboots,trainpreds,traintargs,olsmodel,computed_err_E0[irep]) || !errorvar.efrons_632(nboots,trainpreds,traintargs,olsmodel,computed_err_E632[irep]) ) { Print(" error variance calculation failed "); return; } } PrintFormat("Number of Iterations %d Observed error = %.5lf",ndone, sum_observed_error / double(ndone)) ; //--- PrintFormat("CV: computed error mean=%10.5lf std=%10.5lf",computed_err_cv.Mean(), computed_err_cv.Std()) ; //--- PrintFormat("BOOT: computed error mean=%10.5lf std=%10.5lf",computed_err_boot.Mean(), computed_err_boot.Std()) ; //--- PrintFormat("E0: computed error mean=%10.5lf std=%10.5lf",computed_err_E0.Mean(), computed_err_E0.Std()) ; //--- PrintFormat("E632: computed error mean=%10.5lf std=%10.5lf",computed_err_E632.Mean(), computed_err_E632.Std()) ; } //+--------------------------------------------------------------------+
A scatter plot of the data for a given a 'PredictionDifficultyLevel' of 1.0, would reveal an elliptical distribution with its major axis oriented diagonally upward and to the right. The 'PredictionDifficultyLevel' parameter in the script controls the degree of separation between the clusters, allowing for easier class identification. The lower this parameter is, the harder it will be for the model to infer class membership.
Two distinct classes are generated, with their respective data distributions shifted approximately perpendicular to the major axis by a user-specified magnitude. A linear model is fitted to the data, with the predicted variable assigned a value of −1.0 for one class and +1.0 for the other. The prediction error is defined as 0.0 if the true and predicted values share the same sign, and 1.0 if they exhibit opposite signs. This binary error metric reflects the inherent nature of classification problems.
Two distinct experimental evaluations were conducted. The first experiment utilized a sample size of 15 observations, 1,000 bootstrap repetitions, 100 trials, and a separation of zero. This configuration effectively simulated a scenario with no discriminative information, as the two classes exhibited identical distributions. As anticipated, the observed mean error was around 0.5. The results of this experiment are presented next:
OO 0 10:35:04.051 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) Number of Iterations 100 Observed error = 0.50267 PS 0 10:35:04.051 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) CV: computed error mean= 0.50267 std= 0.18389 KM 0 10:35:04.051 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) BOOT: computed error mean= 0.45214 std= 0.11748 EQ 0 10:35:04.051 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) E0: computed error mean= 0.50517 std= 0.10845 RF 0 10:35:04.051 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) E632: computed error mean= 0.45196 std= 0.09941
Cross-validation once again demonstrated its near-unbiased nature. This result is expected in this scenario, as the model's lack of predictive power results in a 50% probability of misclassification for any given observation. Analogous reasoning applies to the E0 estimator. While E0 is typically expected to exhibit pessimistic bias, this characteristic manifests only when the model possesses some degree of effectiveness. In this instance, E0's forced exclusion of test observations from the training set has a neutral impact.
However, this neutrality does not extend to the E632 estimator. Due to the combination of a truly unbiased component and a strongly optimistically biased component, E632 exhibits significant optimistic bias. This potential bias warrants careful consideration. This experiment also highlights the principal criticism of cross-validation: its high variance. The standard deviation of the cross-validation estimator is substantially higher than that of the E0 estimator. As is frequently observed, the E632 estimator exhibits the lowest standard deviation. However, this advantage is diminished by the strong optimistic bias inherent in E632.
A second experiment, in which the model possessed some predictive capability, was also performed, (the PredictionDifficultyLevel parameter was increased to 1.0). The results of this experiment are presented below:
GM 0 10:38:15.306 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) Number of Iterations 100 Observed error = 0.00747 NM 0 10:38:15.306 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) CV: computed error mean= 0.00533 std= 0.01909 RO 0 10:38:15.306 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) BOOT: computed error mean= 0.00716 std= 0.01766 OG 0 10:38:15.306 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) E0: computed error mean= 0.01012 std= 0.01878 FD 0 10:38:15.306 ErrorVarianceEstimation_ClassificationDemo (Gold RSI Trend Up Index,H1) E632: computed error mean= 0.00820 std= 0.01869
This experiment demonstrates that the E632 estimator is the most effective. It exhibits low bias and the lowest standard deviation. However, the E0 estimator maintains a moderately higher standard deviation and exhibits its characteristic, and often desirable, pessimistic bias. Considering the results of the previous experiment, this factor should be carefully evaluated when selecting between the E0 and E632 estimators. Once more, cross-validation exhibits the highest standard deviation, and the straight bootstrap exhibits a potentially dangerous optimistic bias.
A note on using the algorithms presented for estimating classification performance. There is a significant limitation related to generating bootstrapped classification datasets. Sometimes a bootstrap sample set may end up containing exemplars of a single class, causing the classification algorithm problems or in some instances, to completely fail. Readers who wish to employ the techniques described should be aware of such potential pitfalls.
Conclusion
A cursory examination of the content of this article may lead to the perception that resampling techniques for estimating a model's population error are conceptually intriguing but excessively complex and of limited practical utility. Such a conclusion would be regrettable, as the resampling methodologies presented here offer substantial advantages and warrant serious consideration.
Specifically, these methods address a fundamental challenge in model evaluation: the requirement for an independent dataset. The conventional approach necessitates the acquisition of a separate dataset, a process that is often logistically burdensome and, in some cases, infeasible.
The resampling techniques discussed in this text obviate this requirement. The entire dataset can be utilized for both model training and the estimation of its future performance, thereby maximizing data utilization. This capability represents a significant methodological advancement and should not be dismissed lightly.
| File Name | File Description |
|---|---|
| MQL5/include/error_variance_estimation.mqh | A header file containing definitions of the error estimation algorithms described in the article. |
| MQL5/include/imodel.mqh | The header file includes the definition of interfaces used to interact with machine learning models. |
| MQL5/include/np.mqh | A header file of various vector and matrix utility funtions. |
| MQL5/include/OLS.mqh | The include file defines the OLS class that implements ordinary least squares models. |
| MQL5/scripts/ErrorVarianceEstimation_NumericalPredictionDemo.mq5 | Demonstration script that shows the utility of error estimation algorithms in numerical prediction. |
| MQL5/scripts/ErrorVarianceEstimation_ClassificationDemo.mq5 | Demonstration script that shows the utility of error estimation algorithms in data classification. |
Warning: All rights to these materials are reserved by MetaQuotes Ltd. Copying or reprinting of these materials in whole or in part is prohibited.
This article was written by a user of the site and reflects their personal views. MetaQuotes Ltd is not responsible for the accuracy of the information presented, nor for any consequences resulting from the use of the solutions, strategies or recommendations described.
 Exploring Advanced Machine Learning Techniques on the Darvas Box Breakout Strategy
Exploring Advanced Machine Learning Techniques on the Darvas Box Breakout Strategy
 From Novice to Expert: Support and Resistance Strength Indicator (SRSI)
From Novice to Expert: Support and Resistance Strength Indicator (SRSI)
 Price Action Analysis Toolkit Development (Part 18): Introducing Quarters Theory (III) — Quarters Board
Price Action Analysis Toolkit Development (Part 18): Introducing Quarters Theory (III) — Quarters Board
 USD and EUR index charts — example of a MetaTrader 5 service
USD and EUR index charts — example of a MetaTrader 5 service
- Free trading apps
- Over 8,000 signals for copying
- Economic news for exploring financial markets
You agree to website policy and terms of use