Data Science and Machine Learning (Part 18): The battle of Mastering Market Complexity, Truncated SVD Versus NMF
-- The more you have, the less you see!
Contents:
- What is Dimensionality Reduction?
- High Dimensional Data
- Curse of Dimensionality
- What is Truncated SVD?
- Calculating Explained Variance Function
- Dimensionality Reduction using Truncated SVD
- Advantages of Truncated SVD
- Advantages of NMF
- Disadvantages of Truncated SVD
- Disadvantages of NMF
- Dimensionality Reduction Trade-offs
- FAQs on Dimensionality Reduction
Dimensionality Reduction
According to Wikipedia, Dimensionality Reduction is the transformation of data from a high-dimensional space into a low dimensional space so that the low-dimensional representation retains some meaningful properties of the original data, ideally close to its intrinsic dimension.
Working in high-dimensional spaces can be undesirable for many reasons; raw data are often sparse as a consequence of the curse of dimensionality, and analyzing the data is usually computationally intractable (hard to control or deal with). Dimensionality reduction is common in fields that deal with large numbers of observations and/or large numbers of variables, such as signal processing, speech recognition, neuroinformatics, and bioinformatics read more.
Below are some key points of dimensionality reduction.
High-Dimensional Data
Let's be real for a second, шn most real-world applications many datasets used to build machine-learning models have a very large number of features or variables(dimensions). High-dimensional data can lead to all sorts of challenges such as Increased computation complexity, the risk of overfitting, and difficulties in visualization. That dataset you usually use with 5 independent variables! That's not what the big guys in AI-Algorithmic trading do.
Say you collect all the MT5(38) built-in indicator buffers. You end up with 56 buffers worth of data. This dataset is now huge.
Curse of Dimensionality
This curse is real, and for those who don't believe, try to implement a Linear regression model with a lot of correlated independent variables.The presence of highly correlated features can cause the machine learning models to capture the noise and specific patterns present in the training data, which may not generalize well to new, unseen data.
The curse of dimensionality refers to the problems that arise when dealing with high-dimensional data, such as sparse data distribution, increased computational requirements, and the risk of model overfitting read more.
Using a linear regression model, let me demonstrate this closely:
Since the first 13 indicators in the CSV file are trend-following indicators, they are most likely to be correlated with themselves and with the market despite being of different dimensions.
When only 10 trend-following indicators were used to predict the close prices for 100 bars using our linear regression model:
matrix_utils.TrainTestSplitMatrices(data, train_x, train_y, test_x, test_y); lr.fit(train_x, train_y, NORM_MIN_MAX_SCALER); vector preds = lr.predict(train_x); Print("Train acc = ",metrics.r_squared(train_y, preds));
The result was a 92.2% accurate model on the training phase.
IH 0 16:02:57.760 dimension reduction test (EURUSD,H1) Train acc = 0.9222314640780123
Even the predictions were so accurate.
KE 0 07:10:55.652 GetAllIndicators Data (EURUSD,H1) Actuals GS 0 07:10:55.652 GetAllIndicators Data (EURUSD,H1) [1.100686090182938,1.092937381937508,1.092258894809645,1.092946246629209,1.092958015748051,1.093392517797872,1.091850551839335,1.093040013995282,1.10067748979471,1.09836069904319,1.100275794247118,1.09882067865937,1.098572498565924,1.100543446136921,1.093174625248424,1.092435707204331,1.094128601953777,1.094192935809526,1.100305866167228,1.098452358470866,1.093010702771546,1.098612431849777,1.100827466038129,1.092880150279397,1.092600810699407,1.098722104612313,1.100707460497776,1.09582736814953,1.093765475161778,1.098767398966827,1.099091982657956,1.1006214183736,1.100698195592653,1.092047903797313,1.098661598784805,1.098489471676998,1.0997812203466,1.091954251247136,1.095870956581002,1.09306319770129,1.092915244023817,1.09488564050598,1.094171202526975,1.092523345374652,1.100564904733422,1.094200831112628,1.094001716368673,1.098530017588284,1.094081896433971,1.099230887219729,1.092892028948739,1.093709694144579,1.092862170694582,1.09148709705318,1.098520929394599,1.095105152264984,1.094272325978734,1.098468177450342,1.095849714911251,1.097952718476183,1.100746388049607,1.100114369109941,1.10052138086191,1.096938196194811,1.099992890418429,1.093106549957034,1.095523586088275,1.092801661288758,1.095956895328893,1.100419992807803] PO 0 07:10:55.652 GetAllIndicators Data (EURUSD,H1) Preds DH 0 07:10:55.652 GetAllIndicators Data (EURUSD,H1) [1.10068609018278,1.092937381937492,1.092258894809631,1.09294624662921,1.092958015748061,1.093392517797765,1.091850551839298,1.093040013995258,1.100677489794526,1.098360699043097,1.100275794246983,1.098820678659222,1.09857249856584,1.1005434461368,1.093174625248397,1.09243570720428,1.094128601953754,1.094192935809492,1.100305866167089,1.098452358470756,1.09301070277155,1.098612431849647,1.100827466038003,1.092880150279385,1.092600810699346,1.098722104612175,1.100707460497622,1.095827368149497,1.093765475161723,1.098767398966682,1.099091982657809,1.100621418373435,1.100698195592473,1.092047903797267,1.098661598784675,1.098489471676911,1.099781220346472,1.091954251247108,1.095870956580967,1.09306319770119,1.092915244023811,1.094885640505868,1.094171202526922,1.092523345374596,1.100564904733304,1.094200831112605,1.094001716368644,1.098530017588172,1.094081896433954,1.099230887219588,1.092892028948737,1.093709694144468,1.092862170694582,1.091487097053125,1.098520929394468,1.09510515226494,1.094272325978626,1.098468177450255,1.095849714911142,1.097952718476091,1.100746388049438,1.100114369109807,1.100521380861786,1.096938196194706,1.099992890418299,1.093106549956975,1.09552358608823,1.092801661288665,1.095956895328861,1.100419992807666]
I believe you and I both can agree that this is too good to be true. Despite the model showing an impressive 87.5% accuracy in the testing sample, about 5% accuracy drop.
IJ 0 16:02:57.760 dimension reduction test (EURUSD,H1) Test acc = 0.8758590697252272
We can use the most popular dimension reduction technique called Principal Component Analysis(PCA), to reduce the dimensions of our dataset which now has 10 columns.
//--- Reduce dimension pca = new CPCA(8); //Reduce the dimensions of the data to 8 columns data = pca.fit_transform(data); pca.extract_components(data, CRITERION_SCREE_PLOT);
This resulted in a 88.98% accurate linear regression model during training and also 89.991% during testing, The accuracy didn't drop to say the least.
DJ 0 16:03:23.890 dimension reduction test (EURUSD,H1) Train acc = 0.8898608919375138 RH 0 16:03:23.890 dimension reduction test (EURUSD,H1) Test acc = 0.8991693574000851
Still an overfitted model but a lot better than a 92% accurate model when it comes to the ability to generalize which is crucial that a ML model has to be capable of, you can reduce the dimensions further to see what dimensions of the data gave the best results on out-of-sample predictions.
Motivation for Dimensionality Reduction
- Computational Efficiency: Reducing the number of dimensions can speed up the training and execution of machine learning algorithms.
- Improved Generalization: Dimensionality reduction can help improve a model's ability to generalize to new, unseen data just like we have seen in the linear regression model example.
- Visualization: Makes it easier to visualize and interpret data in lower-dimensional space, aiding human understanding.
What is Truncated SVD?
Truncated Singular Value Decomposition(Truncated SVD) is a dimensional reduction technique, just like other dimensionality reduction techniques, it reduces the dimensions of high-dimensional data while retaining most of the original information. This technique is particularly useful in scenarios where the data has a large number of features, making it difficult to perform efficient computations or visualize the data.
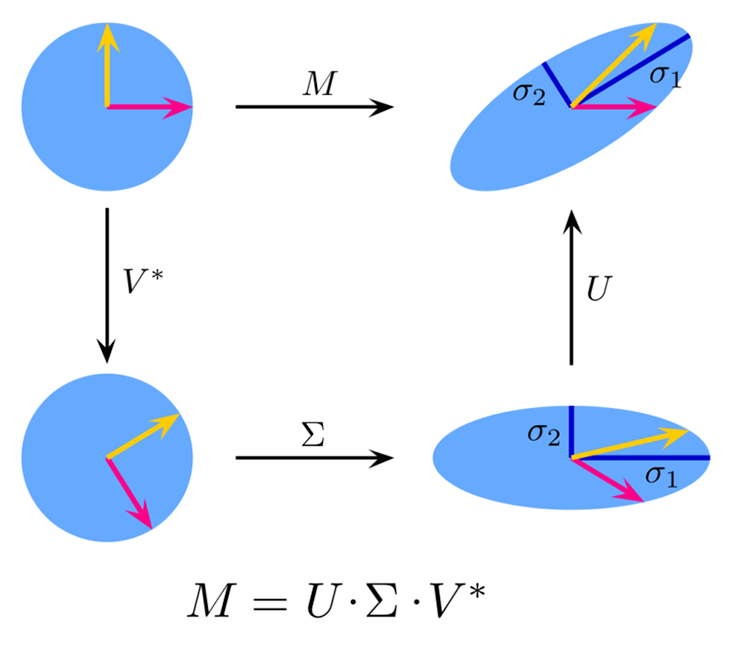
It is a variant of Singular Value Decomposition(SVD) that approximates the original matrix by keeping only the top k singular values and their corresponding singular vectors.
Theory:
image source: Wikipedia
Given a matrix ![]() of dimensions
of dimensions ![]() x
x ![]() , the singular value decomposition of
, the singular value decomposition of ![]() is given by:
is given by:
![]()
where:
![]() is an
is an ![]() ×
× ![]() orthogonal matrix (left singular vectors),
orthogonal matrix (left singular vectors),
![]() is an
is an ![]() ×
× ![]() diagonal matrix with singular values on the diagonal,
diagonal matrix with singular values on the diagonal,
![]() is an
is an ![]() ×
× ![]() orthogonal matrix (right singular vectors).
orthogonal matrix (right singular vectors).
Truncated SVD keeps only the top k singular values and their corresponding vectors:
where:
![]() is an
is an ![]() ×
× ![]() matrix,
matrix,
![]() is a
is a ![]() ×
× ![]() diagonal matrix,
diagonal matrix,
![]() is an
is an ![]() ×
× ![]() matrix.
matrix.
Implementation:
matrix CTruncatedSVD::fit_transform(matrix &X) { n_features = X.Cols(); if (m_components>n_features) { printf("%s Number of dimensions K[%d] is supposed to be <= number of features %d",__FUNCTION__,m_components,n_features); this.m_components = (uint)n_features; } // Center the data (subtract mean) matrix X_centered = CDimensionReductionHelpers::subtract(X, X.Mean(0)); // Compute the covariance matrix matrix cov_matrix = X_centered.Cov(false); // Perform SVD on the covariance matrix matrix U, Vt; vector Sigma; if (!X_centered.SVD(U,Vt,Sigma)) Print(__FUNCTION__," Line ",__LINE__," Failed to calculate SVD Err=",GetLastError()); this.components_ = CDimensionReductionHelpers::Slice(Vt, this.m_components).Transpose(); if (MQLInfoInteger(MQL_DEBUG)) Print("components\n",CDimensionReductionHelpers::Slice(Vt, this.m_components),"\ncomponents_T\n",this.components_); this.explained_variance_ = MathPow(CDimensionReductionHelpers::Slice(Sigma, this.m_components), 2) / (X.Rows() - 1); return X_centered.MatMul(components_); }
There is a built in MQL5 method for calculating SVD of a matrix.
// Singular Value Decomposition. bool matrix::SVD( matrix& U, // unitary matrix matrix& V, // unitary matrix vector& singular_values // singular values vector );
The fit_transform function does the following:
- It centers the data by subtracting the mean.
- It Computes the covariance matrix of the centered data(not important).
- It Performs SVD on the covariance matrix.
- It keeps the top k singular values and vectors.
- It constructs the reduced-dimensional representation of the original matrix.
Calculating Explained Variance Function
The function calculates the total variance by summing the squares of all singular values.
ulong CTruncatedSVD::_select_n_components(vector &singular_values) { double total_variance = MathPow(singular_values.Sum(), 2); vector explained_variance_ratio = MathPow(singular_values, 2).CumSum() / total_variance; if (MQLInfoInteger(MQL_DEBUG)) Print(__FUNCTION__," Explained variance ratio ",explained_variance_ratio); vector k(explained_variance_ratio.Size()); for (uint i=0; i<k.Size(); i++) k[i] = i+1; plt.ScatterCurvePlots("Explained variance plot",k,explained_variance_ratio,"variance","components","Variance"); return explained_variance_ratio.ArgMax() + 1; //Choose k for maximum explained variance }The function takes an array of singular values (singular_values) as input. These singular values are obtained from the Singular Value Decomposition (SVD) of a matrix.
The function then selects the number of components (k) by finding the number of components k with the largest variance. This is achieved using this line of code:
explained_variance_ratio.ArgMax() + 1; We need to modify our fit_transform function to make it capable of detecting the best number of components(when not given any number) to reduce the dimensions of the data and apply the number of components for the truncated SVD.
matrix CTruncatedSVD::fit_transform(matrix &X) { n_features = X.Cols(); if (m_components>n_features) { printf("%s Number of dimensions K[%d] is supposed to be <= number of features %d",__FUNCTION__,m_components,n_features); this.m_components = (uint)n_features; } // Center the data (subtract mean) matrix X_centered = CDimensionReductionHelpers::subtract(X, X.Mean(0)); // Compute the covariance matrix matrix cov_matrix = X_centered.Cov(false); // Perform SVD on the covariance matrix matrix U, Vt; vector Sigma; if (!cov_matrix.SVD(U,Vt,Sigma)) Print(__FUNCTION__," Line ",__LINE__," Failed to calculate SVD Err=",GetLastError()); if (m_components == 0) { m_components = (uint)this._select_n_components(Sigma); Print(__FUNCTION__," Best value of K = ",m_components); } this.components_ = CDimensionReductionHelpers::Slice(Vt, this.m_components).Transpose(); if (MQLInfoInteger(MQL_DEBUG)) Print("components\n",CDimensionReductionHelpers::Slice(Vt, this.m_components),"\ncomponents_T\n",this.components_); this.explained_variance_ = MathPow(CDimensionReductionHelpers::Slice(Sigma, this.m_components), 2) / (X.Rows() - 1); return X_centered.MatMul(components_); }
The default k value is now set to zero to allow this to happen effortlessly.
CTruncatedSVD::CTruncatedSVD(uint k=0) :m_components(k) { }
Dimensionality Reduction using Truncated SVD
Now let's see how the data reduced in dimension by truncated SVD performs:
truncated_svd = new CTruncatedSVD(); data = truncated_svd.fit_transform(data); Print("Reduced matrix\n",data); //--- matrix train_x, test_x; vector train_y, test_y; data = matrix_utils.concatenate(data, target); //add the target variable to the dataset that is either normalized or not matrix_utils.TrainTestSplitMatrices(data, train_x, train_y, test_x, test_y, 0.7, 42); lr.fit(train_x, train_y, NORM_STANDARDIZATION); //training Linear regression model vector preds = lr.predict(train_x); //Predicting the training data Print("Train acc = ",metrics.r_squared(train_y, preds)); //Measuring the performance preds = lr.predict(test_x); //predicting the test data Print("Test acc = ",metrics.r_squared(test_y, preds)); //measuring the performance
Outputs:
LH 0 20:13:41.385 dimension reduction test (EURUSD,H1) CTruncatedSVD::_select_n_components Explained variance ratio [0.2399955100411572,0.3875113031818686,0.3903532427910986,0.3929609228375971,0.3932960517565894,0.3933072531960168,0.3933072531960282,0.3933072531960282,0.3933072531960282,0.3933072531960282] EH 0 20:13:41.406 dimension reduction test (EURUSD,H1) CTruncatedSVD::fit_transform Best value of K = 7 ... ... ... MR 0 20:13:41.407 dimension reduction test (EURUSD,H1) Train acc = 0.8934645199970468 HP 0 20:13:41.407 dimension reduction test (EURUSD,H1) Test acc = 0.8988671205298875
A stunning and stable performance 89.3% accuracy on training while 89.8% accuracy on testing. What about NMF?
What is NMF?
NMF stands for Non-Negative Matrix Factorization, and it is a dimensionality reduction technique that factorizes a matrix into two non-negative matrices. The goal of NMF is to represent the original matrix as the product of two lower-dimensional matrices, where all elements in the matrices are non-negative.
Given an input matrix X of dimensions m×n, NMF factorizes it into two matrices W and H such that:
![]()
Where:
![]() is an
is an ![]() ×
× ![]() matrix, where k is the number of components or features.
matrix, where k is the number of components or features.
![]() is a
is a ![]() ×
× ![]() matrix.
matrix.
Both ![]() and
and ![]() have non-negative entries, and this non-negativity constraint makes NMF particularly suitable for applications where the data is naturally non-negative, such as image data, text data, and spectrograms.
have non-negative entries, and this non-negativity constraint makes NMF particularly suitable for applications where the data is naturally non-negative, such as image data, text data, and spectrograms.
The factorization is achieved by minimizing the Frobenius norm of the difference between the original matrix ![]() and its approximation
and its approximation ![]() ×
× ![]() . Mathematically, this can be expressed as:
. Mathematically, this can be expressed as:
![]()
where ![]() denotes the Frobenius norm.
denotes the Frobenius norm.
Implementation:
Transform Method
matrix CNMF::transform(matrix &X) { n_features = X.Cols(); if (m_components>n_features) { printf("%s Number of dimensions K[%d] is supposed to be <= number of features %d",__FUNCTION__,m_components,n_features); this.m_components = (uint)n_features; } if (this.W.Rows()==0 || this.H.Rows()==0) { Print(__FUNCTION__," Model not fitted. Call fit method first."); matrix mat={}; return mat; } return X.MatMul(this.H.Transpose()); }
The transform function transforms new data X using the already fitted NMF components.
Fit Transform Method
matrix CNMF::fit_transform(matrix &X, uint k=2) { ulong m = X.Rows(), n = X.Cols(); double best_frobenius_norm = DBL_MIN; m_components = m_components == 0 ? (uint)n : k; //--- Initialize Random values this.W = CMatrixutils::Random(0,1, m, this.m_components, this.m_randseed); this.H = CMatrixutils::Random(0,1,this.m_components, n, this.m_randseed); //--- Update factors vector loss(this.m_max_iter); for (uint i=0; i<this.m_max_iter; i++) { // Update W this.W *= MathAbs((X.MatMul(this.H.Transpose())) / (this.W.MatMul(this.H.MatMul(this.H.Transpose()))+ 1e-10)); // Update H this.H *= MathAbs((this.W.Transpose().MatMul(X)) / (this.W.Transpose().MatMul(this.W.MatMul(this.H))+ 1e-10)); loss[i] = MathPow((X - W.MatMul(H)).Flat(1), 2); // Calculate Frobenius norm of the difference double frobenius_norm = (X - W.MatMul(H)).Norm(MATRIX_NORM_FROBENIUS); if (MQLInfoInteger(MQL_DEBUG)) printf("%s [%d/%d] Loss = %.5f frobenius norm %.5f",__FUNCTION__,i+1,m_max_iter,loss[i],frobenius_norm); // Check convergence if (frobenius_norm < this.m_tol) break; } return this.W.MatMul(this.H); }
The fit_transform method performs NMF factorization on the input matrix X and returns the product of matrices W and H.
unlike the truncated SVD which we were able to write code to determine the number of components and apply it to itself, making using the truncated library easier, The NMF learning algorithm is quite different as it requires iterating over an n amount of iterations to find the best values for the algorithm, during the loop the number of k components must remain the same, that's why the fit_transform function takes the value of k components as one of its arguments.
We can get the best number of components for the Non-negative matrix factorization algorithm using the function select_best_components:
uint CNMF::select_best_components(matrix &X) { uint best_components = 1; this.m_components = (uint)X.Cols(); vector explained_ratio(X.Cols()); for (uint k = 1; k <= X.Cols(); k++) { // Calculate explained variance or other criterion matrix X_reduced = fit_transform(X, k); // Calculate explained variance as the ratio of squared Frobenius norms double explained_variance = 1.0 - (X-X_reduced).Norm(MATRIX_NORM_FROBENIUS) / (X.Norm(MATRIX_NORM_FROBENIUS)); if (MQLInfoInteger(MQL_DEBUG)) printf("k %d Explained Var %.5f",k,explained_variance); explained_ratio[k-1] = explained_variance; } return uint(explained_ratio.ArgMax()+1); }
Due to the randomized nature of both Basic and Coefficient matrices, the results of the iterative multiplicative algorithm will be the random values too, so the outcome should be unpredictable unless the random state is set to some value greater than zero.
class CNMF { protected: uint m_components; uint m_max_iter; int m_randseed; ulong n_features; matrix W; //Basic matrix matrix H; //coefficient matrix double m_tol; //loss tolerance public: CNMF(uint max_iter=100, double tol=1e-4, int random_state=-1); ~CNMF(void); matrix fit_transform(matrix &X, uint k=2); matrix transform(matrix &X); uint select_best_components(matrix &X); };
Dimensionality Reduction using NMF
Now let's see how the data reduced in dimension by Non-negative Matrix Factorization performs:
nmf = new CNMF(30); data = nmf.fit_transform(data, 10); Print("Reduced matrix\n",data); //--- matrix train_x, test_x; vector train_y, test_y; data = matrix_utils.concatenate(data, target); //add the target variable to the dataset that is either normalized or not matrix_utils.TrainTestSplitMatrices(data, train_x, train_y, test_x, test_y, 0.7, 42); lr.fit(train_x, train_y, NORM_STANDARDIZATION); //training Linear regression model vector preds = lr.predict(train_x); //Predicting the training data Print("Train acc = ",metrics.r_squared(train_y, preds)); //Measuring the performance preds = lr.predict(test_x); //predicting the test data Print("Test acc = ",metrics.r_squared(test_y, preds)); //measuring the performance
Outcomes:
DG 0 11:51:07.197 dimension reduction test (EURUSD,H1) Best k components = 10 LG 0 11:51:07.197 dimension reduction test (EURUSD,H1) CNMF::fit_transform [1/30] Loss = 187.40949 frobenius norm 141.12462 EG 0 11:51:07.198 dimension reduction test (EURUSD,H1) CNMF::fit_transform [2/30] Loss = 106.49597 frobenius norm 130.94039 KS 0 11:51:07.198 dimension reduction test (EURUSD,H1) CNMF::fit_transform [3/30] Loss = 84.38553 frobenius norm 125.05413 OS 0 11:51:07.198 dimension reduction test (EURUSD,H1) CNMF::fit_transform [4/30] Loss = 67.07345 frobenius norm 118.96900 OR 0 11:51:07.198 dimension reduction test (EURUSD,H1) CNMF::fit_transform [5/30] Loss = 52.50290 frobenius norm 112.46587 LR 0 11:51:07.198 dimension reduction test (EURUSD,H1) CNMF::fit_transform [6/30] Loss = 40.14937 frobenius norm 105.48081 RR 0 11:51:07.198 dimension reduction test (EURUSD,H1) CNMF::fit_transform [7/30] Loss = 29.79307 frobenius norm 98.11626 IR 0 11:51:07.198 dimension reduction test (EURUSD,H1) CNMF::fit_transform [8/30] Loss = 21.32224 frobenius norm 90.63011 NS 0 11:51:07.198 dimension reduction test (EURUSD,H1) CNMF::fit_transform [9/30] Loss = 14.63453 frobenius norm 83.36462 HL 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [10/30] Loss = 9.58168 frobenius norm 76.62838 NL 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [11/30] Loss = 5.95040 frobenius norm 70.60136 DM 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [12/30] Loss = 3.47775 frobenius norm 65.31931 LM 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [13/30] Loss = 1.88772 frobenius norm 60.72185 EN 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [14/30] Loss = 0.92792 frobenius norm 56.71242 RO 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [15/30] Loss = 0.39182 frobenius norm 53.19791 NO 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [16/30] Loss = 0.12468 frobenius norm 50.10411 NH 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [17/30] Loss = 0.01834 frobenius norm 47.37568 KI 0 11:51:07.199 dimension reduction test (EURUSD,H1) CNMF::fit_transform [18/30] Loss = 0.00129 frobenius norm 44.96962 KI 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [19/30] Loss = 0.02849 frobenius norm 42.84850 MJ 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [20/30] Loss = 0.07289 frobenius norm 40.97668 RJ 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [21/30] Loss = 0.11920 frobenius norm 39.31975 QK 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [22/30] Loss = 0.15954 frobenius norm 37.84549 FD 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [23/30] Loss = 0.19054 frobenius norm 36.52515 RD 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [24/30] Loss = 0.21142 frobenius norm 35.33409 KE 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [25/30] Loss = 0.22283 frobenius norm 34.25183 PF 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [26/30] Loss = 0.22607 frobenius norm 33.26170 GF 0 11:51:07.200 dimension reduction test (EURUSD,H1) CNMF::fit_transform [27/30] Loss = 0.22268 frobenius norm 32.35028 NG 0 11:51:07.201 dimension reduction test (EURUSD,H1) CNMF::fit_transform [28/30] Loss = 0.21417 frobenius norm 31.50679 CG 0 11:51:07.201 dimension reduction test (EURUSD,H1) CNMF::fit_transform [29/30] Loss = 0.20196 frobenius norm 30.72260 CP 0 11:51:07.201 dimension reduction test (EURUSD,H1) CNMF::fit_transform [30/30] Loss = 0.18724 frobenius norm 29.99074 ... ... FS 0 11:51:07.201 dimension reduction test (EURUSD,H1) Train acc = 0.604329883526616 IR 0 11:51:07.202 dimension reduction test (EURUSD,H1) Test acc = 0.5115967009955317
The function select_best_components found that the best number of components was 10, the same as the number of columns in the dataset we gave it, It is fair to say that the accuracy of the data resulted from NMF when applied to the linear regression model produced average results of 61% during training and 51% during testing. Before we conclude that TruncatedSVD is a clear winner in this situation However, we need to acknowledge the fact that Non-negative Matrix Factorization(NMF) requires much programming to make it work. When implemented properly and with a better learning algorithm such as Stochastic Gradient Descent(SGD) or any other smart learning algorithm, it can produce a better dataset but for the basic implementations on both algorithms! Truncated SVD won(on paper though)
Let's see how both perform in the big data:
Let's apply the entire data to our algorithms and measure how would they fare in the entire dataset, This will gives us a clue as to which method should be applied to get the best compressed data for our linear regression model.
Truncated SVD Versus NMF, The Battle
This time no data was filtered, we went all in with all the indicators applied for the truncated SVD.
void TrainTestLR(int start_bar=1) { string names; matrix data = indicators.GetAllBuffers(names, start_bar, buffer_size); //--- Getting close values MqlRates rates[]; ArraySetAsSeries(rates, true); CopyRates(Symbol(), PERIOD_CURRENT, start_bar, buffer_size, rates); double targ_Arr[]; ArrayResize(targ_Arr, buffer_size); for (int i=0; i<buffer_size; i++) targ_Arr[i] = rates[i].close; vector target = matrix_utils.ArrayToVector(targ_Arr); //--- Dimension reduction process switch(dimension_redux) { case NMF: { nmf = new CNMF(nmf_iterations); uint k = nmf.select_best_components(data); //Print("Best k components = ",k); data = nmf.fit_transform(data, k); } break; case TRUNC_SVD: truncated_svd = new CTruncatedSVD(); data = truncated_svd.fit_transform(data); break; case None: break; } //--- Print(EnumToString(dimension_redux)," Reduced matrix[",data.Rows(),"x",data.Cols(),"]\n",data); //--- matrix train_x, test_x; vector train_y, test_y; data = matrix_utils.concatenate(data, target); //add the target variable to the dimension reduced data matrix_utils.TrainTestSplitMatrices(data, train_x, train_y, test_x, test_y, 0.7, 42); lr.fit(train_x, train_y, NORM_STANDARDIZATION); //training Linear regression model vector preds = lr.predict(train_x); //Predicting the training data Print("Train acc = ",metrics.r_squared(train_y, preds)); //Measuring the performance preds = lr.predict(test_x); //predicting the test data Print("Test acc = ",metrics.r_squared(test_y, preds)); //measuring the performance }
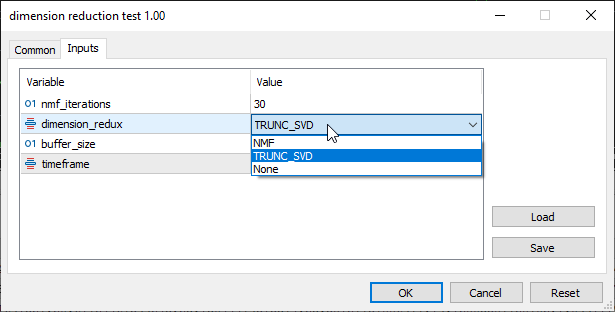
In the inputs section TRUNC_SVD which stands for Truncated SVD was selected:
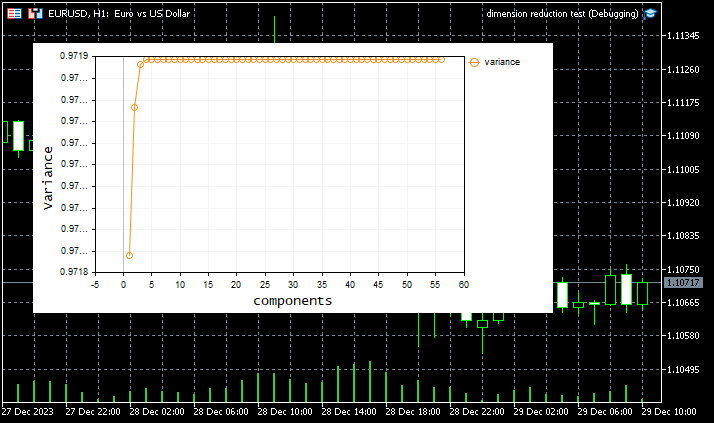
The results were outstanding as expected for truncated SVD. The data was compressed into 11 useful components at the pivotal point of the graph below
Outputs:
KI 0 13:35:07.262 dimension reduction test (EURUSD,H1) [56x56] Covariance HS 0 13:35:07.262 dimension reduction test (EURUSD,H1) [[1.659883222775595e-05,0.03248108864728622,0.002260592127494286,0.002331226815118805,0.0473802958063898,0.009411152185446843,-0.0007144995075063451,1.553267567351765e-05,1.94117385500497e-05,1.165361279698562e-05,1.596183979153003e-05,1.56892102141789e-05,1.565786314082391e-05,1.750827764889262e-05,1.568797564383075e-05,1.542116504856457e-05,1.305965012072059e-05,1.184306318548672e-05,0,1.578309112710041e-05,1.620880130984106e-05,3.200794587841053e-07,1.541769717649654e-05,1.582152318930163e-05,5.986572608120619e-07,-7.489818958341503e-07,-1.347949386573036e-07,1.358668593395908,-0.0551110816439555,7.279528408421348e-05,-0.0001838813666139953,5.310112938097826e-07,6.759105066381161e-07,1.755806692036581e-05,-1.448992128283333e-07,0.003656398187537544,-2.423948560741599e-05,1.65581437719033e-05,-0.01251289868226832,-0.007951606834421109,8.315844054428887e-08,-0.02211745766272421,58.3452835083685,-0.004138620879652527,115.0719348800515,0.7113815226449098,-3.467421230325816e-07,1.456828920113124e-05,1.536603660063518e-05,1.576222466715787e-05,6.85495465028602e-07,0,0,1.264184887739262e-06,-8.035590653384948e-05,-6.948836688267333e-07] DG 0 13:35:07.262 dimension reduction test (EURUSD,H1) [0.03248108864728622,197.2946284096305,47.5593359902222,-70.84984591299812,113.00968468357,80.50832448214619,-85.73100643892406,0.01887146397983594,0.03089371802428824,0.006849209935383654,0.03467670330041064,0.02416916876582018,0.02412087871833598,0.0345064443900023,0.03070601018237932,0.02036445858890056,0.009641324468627534,0.006973716378187099,0,0.02839705603036633,0.02546230398502955,0.000344615196142723,0.04030780408653764,0.02543558488321825,0.0003508232190957864,0.00918131954773614,0.009950700238139187,18501.50888551419,578.0227911644578,1.963532857377992,7.661630153435527,0.006966837168084083,0.004556433245004339,2.562261625497797,0.002410403923079745,118.382177007498,1.555763207775421,1.620640369180333,97.88352837129584,116.7606184094667,0.0006817870698682788,163.5401896749969,108055.4673427991,140.7005993870342,173684.2642498043,3567.96895955429,0.0026091195479834,0.01088219157686945,0.01632411390047346,0.02372834986595739,0.02071124643451263,0,0,0.003953688163493562,-1.360529605248643,-0.04542261766857811] IL 0 13:35:07.262 dimension reduction test (EURUSD,H1) [0.002260592127494286,47.5593359902222,39.19958980374253,-35.49971367926001,7.967839162735706,33.38887983973432,-33.90876164339727,-0.001453063639065681,-0.000297822858867019,-0.002608304419264346,0.002816269112655834,-0.0006711791217058063,-0.0006698381044796457,0.002120545460717355,0.002300146098203005,0.0001826452994732207,-0.0003768483866125172,-0.002445364564315893,0,0.0003665645439668078,-0.003400376924338417,-1.141907378821958e-06,0.006568498854351883,0.0006581929826678372,-0.0005502523415197902,0.00579179204268979,0.006348707171855972,7967.929705662135,452.5996105540225,0.7520153701277594,4.211592829265125,0.001461149548206051,0.0001367863805269901,0.8802824558924612,0.001324363167679062,50.70542641656378,0.6826775813408293,0.5131585731076505,116.3828090044126,97.64469627257866,5.244397974811099e-05,113.6246589165999,13312.60086575102,66.02007474397942,15745.06353439358,3879.735235353455,0.002648988695504766,-0.00244724050922883,-0.00263645588222185,-0.001438073133818628,0.005654239425742305,0,0,-0.000119694862927093,-0.3588916052856576,-0.1059287487094797] EM 0 13:35:07.262 dimension reduction test (EURUSD,H1) [0.002331226815118805,-70.84984591299812,-35.49971367926001,60.69058251292376,1.049456584665883,-42.35184208285295,60.90812761343581,0.008675881760275542,0.008620685402656748,0.008731078117894331,-0.000667294720674837,0.005845080481159768,0.005833401998679988,0.002539790385479495,0.001675796458760079,0.00792229604709134,0.01072137357191657,0.01189054942647922,0,0.003518070782066417,0.00809138574827074,0.0002045106345618818,-0.005848319055742076,0.005549764528898804,0.00103946798619902,-0.008604821519805591,-0.007960056731330659,-10509.62154107038,-607.7276555152829,-1.225906326607001,-5.179537786920108,-0.003656220590983723,-0.001714862736227047,-1.657652278786164,-0.001941357854756676,-80.61630801951124,-1.158614351229071,-1.010395372975761,-137.5151461158883,-129.0146405010356,…] DK 0 13:35:07.264 dimension reduction test (EURUSD,H1) CTruncatedSVD::_select_n_components Explained variance ratio [0.9718079496976207,0.971876424055512,0.9718966593387935,0.971898669685807,0.9718986697080317,0.9718986697081728,0.9718986697082533,0.9718986697082695,0.9718986697082731,0.9718986697082754,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757,0.9718986697082757] JJ 0 13:35:07.282 dimension reduction test (EURUSD,H1) CTruncatedSVD::fit_transform Best value of K = 11 FE 0 13:35:07.282 dimension reduction test (EURUSD,H1) components GP 0 13:35:07.282 dimension reduction test (EURUSD,H1) [[1.187125882554785e-07,1.707109381366902e-07,-2.162493440707256e-08,-3.212808990986057e-07,1.871672229653978e-07,-1.333159242305641e-05,-1.709658791626167e-05,5.106786021196193e-05,2.160157948893384e-05,-1.302989528292599e-06,4.973487699412528e-06,2.354866764130305e-06,0.0001127065022780296,-0.0001141032171988836,-7.674973628685988e-05,4.318893830956814e-06,-8.017085790662858e-05,1.258581118770149e-05,0.001793756756130711,0.0002550931459979332,0.0005154637372606566,0.001801147973070216,0.0001081777898713252,0.1803191282351903,-0.0971245889819613,0.1677985300894154,0.1116153850447159,0.6136670152098842,0.022810600805461,-0.4096528016535661,-0.04163009420605639,-6.077467743293966e-06,-0.01526638316446551,-0.5288520838909793,-0.03965140358106178,-0.1758179670991415,-0.003645717072725773,-0.0587633049540756,0.004124498383003558,-0.1172419838852104,0.06762215235985558,-0.04328191674666172,0.002573659612267772,0.003811049362514162,-0.0659455961610354,-0.02792883463906511,0.008647163802477509,-0.05514899696636823,0.008017364204121686,2.516946811168054e-06,-0.1623798112317545,6.339565837779877e-07,-9.103773865679859e-08,0,0,0] CH 0 13:35:07.282 dimension reduction test (EURUSD,H1) [0.000187434605818864,0.002812180913638008,0.000925532096115269,0.003073325320378009,0.009404433538995808,-0.1105512584754667,-0.2590049789665196,0.4726584971639716,0.1447383637844528,-0.05374370025421956,-0.752572111353205,0.04852191253671465,-0.1154729372340165,-0.2700032006603445,0.1267147455792905,-0.04806661586591762,-0.0004106947202907203,-0.001517506503982104,-0.003502569576390078,-0.002616630851275089,-0.0003258690084512006,-1.313122566421376e-05,-0.0009386837340425192,7.909925585103072e-05,-7.798476572374581e-05,-8.126463184191059e-05,5.556082790448295e-05,-2.687039190421141e-05,-6.540787996573879e-06,1.330764390715927e-05,-4.491635420079414e-05,-8.144156726118825e-06,-8.508538790002811e-07,5.307068574641107e-05,8.482440778162809e-06,-2.9724306932086e-05,2.843845070244019e-05,1.220433778827776e-05,4.206331251218703e-06,1.256512854012554e-05,7.365925693883173e-06,-1.116338606511276e-05,-2.071419560390051e-06,-3.218285979262829e-06,-1.138651025910271e-05,-7.728111919788928e-06,-4.653652137908648e-06,-9.580813709785919e-06,-1.028740118967147e-05,-6.979591632948435e-06,8.116615752437345e-06,1.763108270729631e-06,-4.348070304093866e-07,0,0,0] JR 0 13:35:07.282 dimension reduction test (EURUSD,H1) [1.848792316776081e-05,0.0008588118715707877,0.001008311173244842,0.002376194713912998,0.03118554398340165,0.07889519945845663,-0.07568381470451135,0.0286506682781276,-0.09873220604411656,-0.09161678196011247,-0.06574722506621368,-0.8477734032486133,-0.2533539865405071,0.01738574140899809,-0.2520128661222155,0.338533576002899,0.03018091859501061,0.03802772499047165,-0.001646103636389454,-0.00230341248842314,0.007310099460075021,-0.006012419384786831,-0.001413152018214914,2.142238614111051e-05,2.944210407515316e-05,-6.521097350129076e-05,4.95794487446122e-05,-8.706533732190571e-06,3.322659358695792e-05,-8.146689287415223e-07,7.338661273889888e-06,6.605547217087068e-06,9.447199984574336e-06,-5.17902002055696e-05,2.513872334761899e-06,-2.256988836110105e-05,-1.911456753589991e-05,-3.264899090321399e-05,-5.800966034108816e-06,1.84157480007614e-05,5.356185659483905e-06,-1.722849771456492e-05,1.407694486288703e-05,-7.853816348797691e-06,-1.555432680785718e-05,1.323568644672602e-05,-6.760913172750632e-06,-1.302135272406512e-05,-3.211793238456227e-06,5.802687671843603e-07,-4.280158053673846e-06,1.549960937140237e-06,-4.919157307518426e-07,0,0,0] ND 0 13:35:07.282 dimension reduction test (EURUSD,H1) [4.113935038265459e-05,-0.00163116425762962,-0.0006793200544509552,-0.002550542034446345,-0.0360619764220519,-0.05083503039040507,0.1457232134777539,-0.1319534223026893,-0.1000465688426969,0.03066773678926198,0.211126338548004,0.02610057183969087,-0.4621259104920466,-0.5881871797892803,0.5329599008633419,0.2326293789423069,-0.004192622153062945,-0.05839467173045784,0.002320319216461524,0.007699098543144081,-0.007658991162669676,-0.003166005488782055,-0.002283364895801648,0.0001978857108759482,9.652410400630472e-05,5.391765247424044e-05,-2.114027852314159e-05,-3.571496353187668e-06,6.919610375993553e-06,-2.388732546126286e-05,-1.795211964869842e-05,-4.375012411627235e-05,-6.279526099805299e-05,2.71361750709131e-06,-2.519904126359203e-05,-3.154274545518064e-05,-4.95034356619025e-05,4.982607182957078e-05,2.91223664932767e-05,7.266298957499022e-06,-5.27732883601878e-0…] QD 0 13:35:07.283 dimension reduction test (EURUSD,H1) TRUNC_SVD Reduced matrix[100x11] FJ 0 13:35:07.283 dimension reduction test (EURUSD,H1) [[3032.510155721446,0.6797958296629614,0.3147476036369294,-1.879105115460979,2.342311343896795,-1.444862488231156,1.72748820545895,9003.45453268309,6583.0037062413,11248.15180107277,7664.89601210886] QF 0 13:35:07.283 dimension reduction test (EURUSD,H1) [2613.746957390415,0.8994353272147151,0.2278876639409569,-1.833173521667439,2.422820504229084,-1.194855144516288,1.619766862896731,8730.428168136412,6321.389560496657,10843.26327739175,7319.502533761807] PE 0 13:35:07.283 dimension reduction test (EURUSD,H1) [2290.391536977857,0.8379176898463352,0.123448650655239,-1.79572777997295,2.553555290053082,-1.261720012039457,1.872612496522998,9359.431555011421,6628.023623123124,11573.90347441899,7725.564567458286] MG 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1793.638587775052,0.8840827856664026,-0.009259140368535656,-1.812929578463889,2.752934946133839,-1.194132624145421,2.137669035967074,10045.24774931484,6958.660329797975,12319.85906895874,8090.242616080116] IG 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1067.248927502046,0.9361386848947133,0.2824462001532311,-2.094258230827017,2.302295904768048,-0.9859566963069195,1.849760818059724,8852.426567434784,5936.081092995532,10850.91447636461,7048.244994007338] NK 0 13:35:07.283 dimension reduction test (EURUSD,H1) [875.831228770905,1.10630868463423,0.3875078994082357,-2.07189694520072,2.003365098290179,-0.8110375102176066,1.363090061871958,7086.981018896034,4689.12320342304,8755.082428498208,5718.774787652458] GK 0 13:35:07.283 dimension reduction test (EURUSD,H1) [750.5105412678478,1.58876151742295,0.4277968820488219,-2.495434492237099,2.546292376170158,-0.5828893757361142,1.468026541982692,8457.543559890122,5579.75159746014,10352.91442474459,6691.476688344664] EK 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1028.864261893481,1.737564309663063,0.6859148896559381,-2.576387294650619,2.22158096752311,-0.3483142589026745,0.9424367352835165,7447.770506698205,5028.271311988544,9145.898159310427,5970.749558054185] MJ 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1315.206321750198,1.599230058650526,1.006274380327883,-2.7208483981554,1.702262719567507,-0.3336344655640055,0.5851633683692923,6123.681144041253,4208.879528858515,7625.280231352584,5091.190116446857] PI 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1040.359603039049,2.015870643904685,1.297584348509677,-3.422661750068991,2.051984203691556,-0.1752179554187868,0.625382499042994,7446.846626605789,4973.286352001192,9200.911151239434,6040.469516939555] MJ 0 13:35:07.283 dimension reduction test (EURUSD,H1) [2056.827104147074,0.6969653180001977,0.5026500071613408,-1.539526411095017,1.333168361525386,-1.205142789226661,0.6457357287148752,4362.936467349854,3118.514061447938,5739.541487386377,4094.4444510553] KI 0 13:35:07.283 dimension reduction test (EURUSD,H1) [2595.720157415741,0.5163475309230526,0.7707831677360033,-1.609464385287863,0.6391376890497206,-1.091660522613616,0.2434028653397404,2245.471581325966,1932.755290423922,3219.551710692477,2600.234934440349] FJ 0 13:35:07.283 dimension reduction test (EURUSD,H1) [2000.813854424802,-0.1645196557230545,0.0271698872028618,-1.136148599089293,1.199419424319289,-1.596267278271581,1.616290624329739,4340.908808342276,3155.426978423527,5643.263654816839,4006.305187235153] GN 0 13:35:07.283 dimension reduction test (EURUSD,H1) [714.201770611258,0.3322650152566928,-0.128924730742915,-1.497052055771412,1.889908667964611,-1.18328154703613,2.040965402442796,6411.664479403819,4214.549479233159,7917.864243044486,5153.2871400763] PN 0 13:35:07.283 dimension reduction test (EURUSD,H1) [676.7845833423586,0.1119963611895549,-0.4614956061196153,-0.7783442013329294,1.680735950870798,-1.110059082233565,1.926793017792492,5379.745050208609,3591.380424598063,6620.153862582291,4307.434978894435] GL 0 13:35:07.283 dimension reduction test (EURUSD,H1) [766.9657214036708,0.3283061830008722,-0.3885600107901345,-1.031650548898253,1.912288786544761,-1.036499883901753,2.01321049032323,6312.725946828255,4249.957068368147,7723.98867795246,5012.0279192467] LN 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1272.697206108888,-0.3546159778199272,-0.7521309963906442,-0.1806331494361637,1.527697751794935,-1.486437217294949,2.136800392269752,5073.89079322631,3559.651506707118,6317.965379534476,4229.397904810504] RM 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1869.479624807111,-0.0175694426730242,-0.7678796689175619,-0.2643173159395689,1.862033543014759,-1.44967470564508,2.028361828380722,5639.625182760593,4124.447639836587,7033.466549718048,4777.911629224665] GN 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1936.02506453734,0.0845940044872068,-0.8282752540396139,-0.2824304083282199,1.982789807497455,-1.350085518779075,2.085883725566998,5911.195073473721,4372.189044548059,7316.534104301676,4945.624347822407] CR 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1844.238763352675,-0.4606751585910102,-1.187448989349241,0.2320045292554417,1.900279565663612,-1.62110507361112,2.534343850099422,5907.730651516376,4364.59879103891,7282.962351393813,4893.24121551531] ER 0 13:35:07.283 dimension reduction test (EURUSD,H1) [1716.303347926322,-0.2450617860724134,-0.9482067759826802,-0.03979477150523128,1.79416546386642,-1.453641927846228,2.284520496569503,5710.722665215339,4183.828568899002,7048.670774203967,4734.907606214866…] GP 0 13:35:07.283 dimension reduction test (EURUSD,H1) Train acc = 0.8087284075835584 ER 0 13:35:07.283 dimension reduction test (EURUSD,H1) Test acc = 0.7286157353628964
80% accurate on the training sample while being 72% accurate on the testing sample.
Non-negative Matrix Factorization(NMF)
On the other hand, the NMF was unable to produce anything of value as anything resulting from it was a nan value, at this point, it is no longer a competitor.
LK 0 13:43:09.997 dimension reduction test (EURUSD,H1) CNMF::fit_transform [1/100] Loss = nan frobenius norm nan II 0 13:43:09.997 dimension reduction test (EURUSD,H1) CNMF::fit_transform [2/100] Loss = nan frobenius norm nan FG 0 13:43:09.997 dimension reduction test (EURUSD,H1) CNMF::fit_transform [3/100] Loss = nan frobenius norm nan KF 0 13:43:09.997 dimension reduction test (EURUSD,H1) CNMF::fit_transform [4/100] Loss = nan frobenius norm nan PD 0 13:43:09.997 dimension reduction test (EURUSD,H1) CNMF::fit_transform [5/100] Loss = nan frobenius norm nan PR 0 13:43:09.998 dimension reduction test (EURUSD,H1) CNMF::fit_transform [6/100] Loss = nan frobenius norm nan KP 0 13:43:09.998 dimension reduction test (EURUSD,H1) CNMF::fit_transform [7/100] Loss = nan frobenius norm nan NO 0 13:43:09.998 dimension reduction test (EURUSD,H1) CNMF::fit_transform [8/100] Loss = nan frobenius norm nan QM 0 13:43:09.998 dimension reduction test (EURUSD,H1) CNMF::fit_transform [9/100] Loss = nan frobenius norm nan MH 0 13:43:09.998 dimension reduction test (EURUSD,H1) CNMF::fit_transform [10/100] Loss = nan frobenius norm nan .... .... LM 0 13:43:10.006 dimension reduction test (EURUSD,H1) CNMF::fit_transform [97/100] Loss = nan frobenius norm nan OO 0 13:43:10.006 dimension reduction test (EURUSD,H1) CNMF::fit_transform [98/100] Loss = nan frobenius norm nan NI 0 13:43:10.006 dimension reduction test (EURUSD,H1) CNMF::fit_transform [99/100] Loss = nan frobenius norm nan CI 0 13:43:10.006 dimension reduction test (EURUSD,H1) CNMF::fit_transform [100/100] Loss = nan frobenius norm nan MI 0 13:43:10.008 dimension reduction test (EURUSD,H1) NMF Reduced matrix[100x56] FN 0 13:43:10.008 dimension reduction test (EURUSD,H1) [[nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan] EF 0 13:43:10.008 dimension reduction test (EURUSD,H1) [nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan] GK 0 13:43:10.008 dimension reduction test (EURUSD,H1) [nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan] QO 0 13:43:10.008 dimension reduction test (EURUSD,H1) [nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan] CL 0 13:43:10.008 dimension reduction test (EURUSD,H1) ... .... EO 0 13:43:10.008 dimension reduction test (EURUSD,H1) [nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan,nan…] EL 0 13:43:10.010 dimension reduction test (EURUSD,H1) Train acc = nan GJ 0 13:43:10.010 dimension reduction test (EURUSD,H1) Test acc = nan
Let's try to build a linear regression model without truncated SVD, let's just apply the entire data to it and observe how its performance.
GM 0 13:53:03.686 dimension reduction test (EURUSD,H1) None Reduced matrix[100x56] MG 0 13:53:03.686 dimension reduction test (EURUSD,H1) [[1.099900334222465,28.09636760954466,23.62682944559725,20.05254468697356,24.9034642580335,25.12500390358301,19.04617822178984,1.098553499999958,1.102834674021181,1.094272325978734,1.100734208567449,1.100855469999989,1.098655958571417,1.100133799959087,1.09983,1.097395,1.0950725,1.094895,1.10265,1.100030000000004,1.09944087984,0.03659425270713104,1.100626095746222,1.098825209694279,0.001265714285714323,1.705459946510501e-05,0.0009170545994650059,1655.083284093067,24.54629344924639,0.4388379204892836,0.1040346153847089,0.001226921314023466,0.0012463900886654,100.1775875415509,-1.946877464193406e-05,63.53135141907871,0.1040413928121892,0.1171616554839711,43.66576819407196,56.26437671196908,0.000170982627085528,-39.78494623655773,-2966278.270646845,59.66122501825306,-426422,2459,-0.0002127529411762598,1.096805768175659,1.098423909197834,1.099767192241293,0.00330055882353042,1.797693134862316e+308,1.797693134862316e+308,0.001618141022174502,0,0.03660024400162265] DH 0 13:53:03.687 dimension reduction test (EURUSD,H1) [1.099895090784212,25.44125240854614,20.47658551951762,17.37887206204375,24.10764337690182,24.31185250851098,18.42976333679036,1.098807999999958,1.102730359493936,1.09488564050598,1.100648597660533,1.100904089999989,1.09870448142856,1.100085345856641,1.09983,1.097395,1.0949775,1.09511,1.10237,1.100145000000004,1.09954,0.03659633771631932,1.100539015261062,1.098860393123325,0.001167857142857174,0.000137475370969975,0.0006774753709699599,1151.094538712408,-2.654176423441059,0.4388379204892836,0.1582615384614039,0.001148578761165142,0.001262448000841409,100.0618688017469,-0.0001138692396762668,61.34588689321424,0.1137518684603838,0.1082289779011129,39.39899833055219,47.44710420521373,0.000170240277350424,-32.25806451612595,-2966207.715091289,53.52331983100873,-424517,1905,-0.0003920588235292355,1.097045324469839,1.098718420548105,1.099909753793035,0.003008823529412785,1.797693134862316e+308,1.797693134862316e+308,0.001673096078265379,0,0.0283464566929126] LG 0 13:53:03.687 dimension reduction test (EURUSD,H1) [1.099896454740888,23.0468607955726,17.74637411691527,20.61724467599245,23.13044479515374,23.58127757886977,19.12803587728702,1.099111999999958,1.102374285088665,1.095849714911251,1.100636052620929,1.100962719999989,1.098762994285703,1.100079584777576,1.09983,1.097395,1.0949025,1.09511,1.10224,1.100252000000004,1.10126,0.03659849012444888,1.10032475011848,1.098869304036258,0.001062857142857168,-6.787825345444531e-05,0.0004021217465455251,385.6853260090575,21.57900929644156,0.4735973597359621,0.1358284615387142,0.001095648513071756,0.001261794377734486,100.0745976729165,-0.00016614586466273,63.02688593246703,0.1054308010829696,0.1073759442661703,21.12676056338241,34.73050902933566,0.000167907541851923,-45.79710144927351,-2967636.417218948,63.24364017027852,-427004,2487,-0.0004969529411762387,1.097296453356775,1.098949867979591,1.099885803034428,0.002709058823530563,1.797693134862316e+308,1.797693134862316e+308,0.001653414622816651,1,0.0188982710092469] OH 0 13:53:03.687 dimension reduction test (EURUSD,H1) [1.09988711983821,22.27698129412946,15.38019090132657,21.80270494870316,22.00511532835773,22.64977536234809,19.53807060936342,1.099376999999958,1.101815803805105,1.096938196194811,1.100524122300143,1.101012769999989,1.098812944285703,1.100014455800224,1.099825,1.097395,1.09474,1.09511,1.10151,1.100228000000004,1.1012264,0.03660061924403133,1.100053014395182,1.098898955035223,0.0009950000000000245,-0.0002124670743897106,0.0003975329256102889,-665.9989270069636,-29.795012402167,0.4599358974358811,0.1292584615385153,0.001012177253020718,0.001241318811166551,100.0636954266684,-0.000229141558145832,59.22581938383797,0.05654610290371272,0.09982547246710144,18.13186813187058,26.21920900860187,0.0001634553208976355,-53.91304347826081,-2969847.417218948,53.21227207754287,-429215,2211,-0.000519241176470371,1.097569033867792,1.099141134482142,1.099908642427543,0.002445970588236479,1.797693134862316e+308,1.797693134862316e+308,0.001572100614350314,1,0.02758932609678876] HJ 0 13:53:03.687 dimension reduction test (EURUSD,H1) [1.099803229128606,19.89136498265082,20.62876885414336,18.89567762220941,21.68993269258447,25.44873831711815,17.83416777391867,1.099577499999958,1.101202281523733,1.097952718476183,…] EK 0 13:53:03.689 dimension reduction test (EURUSD,H1) Train acc = -nan MR 0 13:53:03.689 dimension reduction test (EURUSD,H1) Test acc = -nan
The linear regression model was unable to produce results on its own for this massive dataset, There are some measures that can be taken to address the issues these dimension reduction techniques face such as Normalizing the inputs data, Fixing numerical instability and much more we will discuss in the next articles. You can always track the updates and future changes to these libraries discussed in this article on this GitHub repo:
Let's look at the advantages and disadvantages each Dimension reduction technique offers:
Advantages of Truncated Singular Value Decomposition (SVD)
Preservation of Variance:
SVD aims to preserve as much variance as possible in the original data. It provides a way to represent the data in a lower-dimensional space while retaining the most important information.
Orthogonality of Components:
The components (singular vectors) obtained through SVD are orthogonal, which can be beneficial in certain applications. Orthogonality simplifies the interpretation of components.
Numerical Stability:
SVD is known for its numerical stability. It can handle sparse and noisy data well. The decomposition is based on the eigenvalue decomposition of the covariance matrix.
Applications in Image Compression and Denoising:
SVD is commonly used in image compression and denoising. The singular values provide a measure of importance, and truncating them allows for compression while minimizing loss of information.
Advantages of Non-Negative Matrix Factorization (NMF)
Parts-Based Representation:
NMF enforces non-negativity constraints on both the basis matrix and the coefficient matrix. This leads to a parts-based representation, which can be beneficial in scenarios where parts-based features are meaningful (e.g., in topic modeling or image analysis).
Interpretability:
The non-negativity constraint often results in more interpretable factors. In topics extracted from text data, for example, each topic is represented as a non-negative linear combination of words, making the topics more interpretable.
Applications in Document Analysis and Image Processing:
NMF is commonly used in document analysis (topic modeling) and image processing. It can identify underlying patterns in data and is particularly useful when the data naturally has non-negative components.
Sparse Representations:
NMF tends to produce sparse representations, where only a subset of components is used to represent each data point. This can be advantageous when dealing with high-dimensional data.
Common Advantages
Dimensionality Reduction:
Both SVD and NMF provide effective techniques for reducing the dimensionality of data, which can be crucial for handling high-dimensional datasets and extracting meaningful features.
Feature Extraction:
They can be used for feature extraction, allowing the identification of important patterns or features in the data.
Noise Reduction:
Both techniques can help in denoising data by capturing the essential information and filtering out noise.
Disadvantages of Truncated Singular Value Decomposition (SVD)
Sensitive to Outliers:
SVD can be sensitive to outliers in the data, as it seeks to minimize the sum of squared errors. Outliers can have a significant impact on the decomposition.
Computational Complexity:
The computation of the full SVD can be computationally expensive, especially for large datasets. Truncated SVD provides an approximation but may still be demanding for high-dimensional data.
Limited Applicability to Sparse Data:
SVD may not be as effective when dealing with highly sparse data, as the presence of many zero values can affect the accuracy of the decomposition.
Lack of Interpretability:
While the components obtained from SVD are orthogonal, the interpretability of these components may be challenging, particularly in comparison to NMF, which enforces non-negativity constraints.
Disadvantages of Non-Negative Matrix Factorization (NMF)
Non-Uniqueness of Solutions:
NMF solutions are not unique, meaning that different initializations may lead to different factorizations. This lack of uniqueness can make it challenging to compare results across different runs.
Difficulty with Zero Entries:
NMF may struggle when faced with zero entries in the data matrix. Dealing with missing data or zero values requires special consideration such as removing Nan and Infinity values.
void CDimensionReductionHelpers::ReplaceNaN(matrix &mat) { for (ulong i = 0; i < mat.Rows(); i++) for (ulong j = 0; j < mat.Cols(); j++) if (!MathIsValidNumber(mat[i][j])) mat[i][j] = 0.0; }
Choice of Rank (Number of Components):
Determining the appropriate rank (number of components) for NMF can be challenging. An inappropriate choice may result in overfitting or underfitting the data.
Limited Applicability to Negative Values:
NMF enforces non-negativity, which may not be suitable for datasets where negative values are meaningful. This constraint restricts the types of data that can be effectively modeled.
Common Considerations
Interpretability vs. Reconstruction Accuracy:
NMF may provide more interpretable components, but this often comes at the cost of reconstruction accuracy compared to SVD. The choice between interpretability and accuracy depends on the specific goals of the analysis.
Dimensionality Reduction Trade-offs
Both techniques involve a trade-off between the reduction in dimensionality and the loss of information. The challenge is to find an optimal balance that preserves essential features while minimizing information loss.
Algorithmic Hyperparameters:
Both SVD and NMF may require tuning hyperparameters, such as the number of components or the regularization strength, to achieve optimal results. The sensitivity to these hyperparameters should be considered.
FAQs on Dimensionality Reduction
Question: How does Dimensionality Reduction techniques work?
Answer: They transform the data into a new set of uncorrelated variables components that capture the maximum variance in the original data. They identify the directions along which the data varies the most.
Question: When should I use dimensionality reduction?
Answer: Dimensionality reduction is useful when working with high-dimensional data, such as in machine learning, image processing, and bioinformatics. It is beneficial when there are redundant or irrelevant features, and computational efficiency, interpretability, or visualization is a priority.
Question: Can dimensionality reduction be applied to any type of data
Answer: Dimensionality reduction can be applied to various types of data, including numerical, categorical, and text data. However, the choice of technique and considerations may vary based on the nature of the data.
Question: Are there alternatives to linear dimensionality reduction techniques?
Answer: Yes, there are nonlinear techniques such as Kernel PCA, t-SNE, and autoencoders that capture complex relationships in the data beyond linear transformations.
Question: Is dimensionality reduction always beneficial in machine learning?
Answer: Not necessarily. While dimensionality reduction can offer benefits in terms of computational efficiency, interpretability, and visualization, it may not always improve model performance. It depends on the specific characteristics of the dataset and the machine-learning task.
Question: Can dimensionality reduction cause information loss?
Answer: Yes, dimensionality reduction can lead to information loss, especially if the reduced representation discards relevant features. The goal is to strike a balance between dimensionality reduction and retaining essential information.
Question: What are some challenges of dimensionality reduction techniques?
Answer: Challenges include the potential for overfitting, sensitivity to outliers, the non-uniqueness of solutions (especially in NMF), and the need for careful tuning of hyperparameters.
Attachments:
| File | Description/Usage |
|---|---|
| helpers.mqh | Contains code to help with dimensional reduction operations taking place in both NMF.mqh and TruncatedSVD.mqh |
| Linear Regression.mqh | Contains code for the linear regression model |
| matrix_utils.mqh | Additional matrix operations |
| metrics.mqh | A library containing code for measuring performance of machine learning models |
| NMF | Contains Non-negative Matrix Factorization class |
| plots.mqh | A library for drawing plots on the current chart |
| preprocessing.mqh | A class containing functions for preprocessing data to make it suitable for machine learning |
| TruncatedSVD.mqh | Contains Truncated SVD class |
| dimension reduction test.mq5 | A script for testing all the code discussed in the article |
Peace out.
Warning: All rights to these materials are reserved by MetaQuotes Ltd. Copying or reprinting of these materials in whole or in part is prohibited.
This article was written by a user of the site and reflects their personal views. MetaQuotes Ltd is not responsible for the accuracy of the information presented, nor for any consequences resulting from the use of the solutions, strategies or recommendations described.
 Introduction to MQL5 (Part 2): Navigating Predefined Variables, Common Functions, and Control Flow Statements
Introduction to MQL5 (Part 2): Navigating Predefined Variables, Common Functions, and Control Flow Statements
 MQL5 Wizard Techniques you should know (Part 10). The Unconventional RBM
MQL5 Wizard Techniques you should know (Part 10). The Unconventional RBM
 Developing an MQTT client for Metatrader 5: a TDD approach — Part 5
Developing an MQTT client for Metatrader 5: a TDD approach — Part 5
- Free trading apps
- Over 8,000 signals for copying
- Economic news for exploring financial markets
You agree to website policy and terms of use