Lets open up ms paint and draw a cluster .
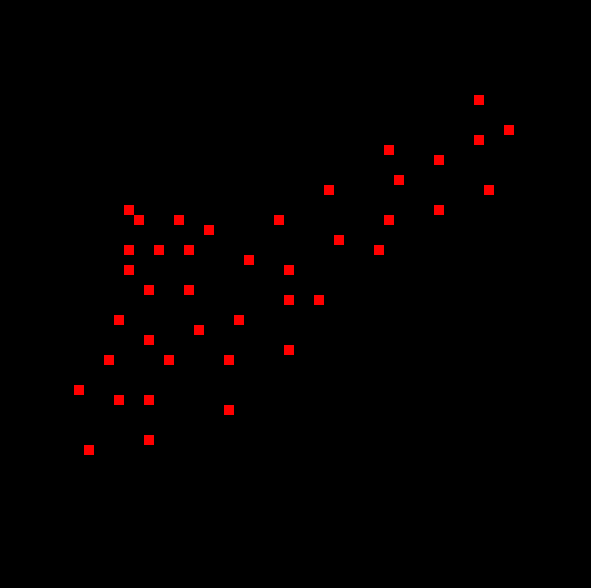
This is an upscale image of a 60x60 manually drawn bmp file
This file will be fed into our code and the code will have to transform it so that the
direction with the biggest variance sits on the x axis (if i understood things right).
So , some preliminary stuff , open the file , resource it , read the pixels , turn them to samples.
Our samples will look like this , we have one class , no fancy stuff.
class a_sample{ public: int x,y,cluster; double xd,yd; a_sample(void){x=0;y=0;cluster=0;} };
Ignore the cluster reading it was meant for another test -which failed-
These are the inputs we'll be working with :
#include <Canvas\Canvas.mqh>; input string testBMP="test_2.bmp";//test file input color fillColor=C'255,0,0';//fill color input color BaseColor=clrWheat;//base color input bool RemoveCovariance=false;//remove covariance on rescale ?
We read the image and turn each pixel that is red (or that matches our color input) to a sample.
//load if(ResourceCreate("TESTRESOURCE","\\Files\\"+testBMP)){ uint pixels[],width,height; if(ResourceReadImage("::TESTRESOURCE",pixels,width,height)){ //collect samples int total_samples=0; uint capture=ColorToARGB(fillColor,255); ArrayResize(samples,width*height,0); //scan the image originalX=(int)width; originalY=(int)height; int co=-1; for(int y=0;y<(int)height;y++){ for(int x=0;x<(int)width;x++){ co++; if(pixels[co]==capture){ total_samples++; samples[total_samples-1].x=x; samples[total_samples-1].y=y; samples[total_samples-1].xd=x; samples[total_samples-1].yd=y; } } } ArrayResize(samples,total_samples,0); Print("Found "+IntegerToString(total_samples)+" samples"); } }
Neat , now we pass the samples into a matrix that is scaled with each sample being a row and each feature (x,y) being a column
matrix original; original.Init(total_samples,2); //fill for(int i=0;i<total_samples;i++){ original[i][0]=samples[i].xd; original[i][1]=samples[i].yd; }
So we shape (resize) the matrix to have total samples # of rows and 2 columns (x , y) . X+Y are our sample features in this example.
We then construct , well not we but the mq library costructs the covariance matrix , we pass false in the parameter as our features sit on the columns . Where our features on the rows , i.e. one row was one feature then we'd call this function using true.
matrix covariance_matrix=original.Cov(false);
What is the covariance matrix ? it is a features by features (features x features) (2x2) matrix that measures the covariance of each feature to all other features . Neat that we can use one line of code .
Then we need the eigenvectors and eigenvalues of that 2x2 covariance matrix
The theory i don't quite understands states that this will indicate the "direction" of most "variance".
Let's play along
matrix eigenvectors; vector eigenvalues; if(covariance_matrix.Eig(eigenvectors,eigenvalues)){ Print("Eigenvectors"); Print(eigenvectors); Print("Eigenvalues"); Print(eigenvalues); }else{Print("Can't Eigen");}
the eigenvectors will be a 2x2 matrix as well if i'm not mistaken , same as the covariance martix.
Now , let's take our samples and "turn" them so that the most varying direction sits on the x axis.If that's how it works and it was not random.
Visualize what we are expecting to see here based on the first image posted.
This is how , so far :
//apply for(int i=0;i<total_samples;i++){ vector thissample={samples[i].xd,samples[i].yd}; thissample=thissample.MatMul(eigenvectors); samples[i].xd=thissample[0]; samples[i].yd=thissample[1]; }
I'm constructing a vector that is one sample , so 2 elements then matrix multiply it with the eigenvectors matrix then pass the resulting x and y back to the samples .
Then some transformation again , the reason is there is to accommodate multiple eigenvector passes , you probably will get the same idea.
//reconstruct double minX=INT_MAX,maxX=INT_MIN,minY=INT_MAX,maxY=INT_MIN; for(int i=0;i<total_samples;i++){ if(samples[i].xd>maxX){maxX=samples[i].xd;} if(samples[i].xd<minX){minX=samples[i].xd;} if(samples[i].yd>maxY){maxY=samples[i].yd;} if(samples[i].yd<minY){minY=samples[i].yd;} } //0 to 1 double rangeX=maxX-minX; double rangeY=maxY-minY; double allMax=MathMax(maxX,maxY); double allMin=MathMin(minX,minY); double allRange=allMax-allMin; for(int i=0;i<total_samples;i++){ samples[i].xd=((samples[i].xd-minX)/rangeX)*1000.0; samples[i].yd=((samples[i].yd-minY)/rangeY)*1000.0; samples[i].x=(int)samples[i].xd; samples[i].y=(int)samples[i].yd; } originalX=1000; originalY=1000;
Then the drawing part which is not that important .
this is the outcome
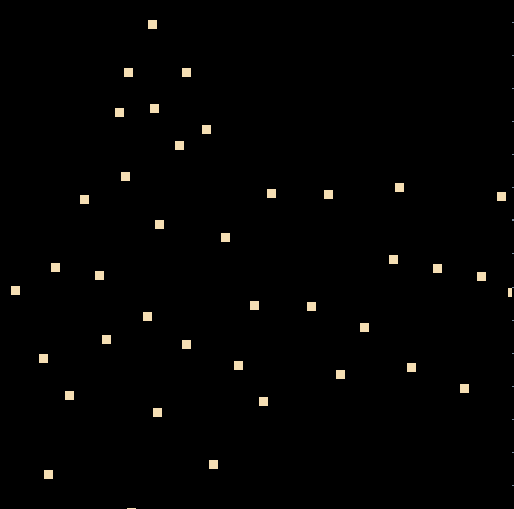
The samples touching the edges is more due to our reconstruction but the transformation occured.
Here is the full code , i've left my mistakes in commented out , and also the removeCovariance is not working.
If you find issues let me know.
Now imagine you have 1000 features right ? you cannot plot all that so how do we construct a new feature that is a synthetic weighted sum of the old features based on the eigenvectors (if i'm on the right track here) . Like Principle component analysis .
#property version "1.00" #include <Canvas\Canvas.mqh>; input string testBMP="test_2.bmp";//test file input color fillColor=C'255,0,0';//fill color input color BaseColor=clrWheat;//base color input bool RemoveCovariance=false;//remove covariance on rescale ? string system_tag="TEST_"; class a_sample{ public: int x,y,cluster; double xd,yd; a_sample(void){x=0;y=0;cluster=0;} }; int DISPLAY_X,DISPLAY_Y,originalX,originalY; double PIXEL_RATIO_X=1.0,PIXEL_RATIO_Y=1.0; bool READY=false; a_sample samples[]; CCanvas DISPLAY; int OnInit() { ArrayFree(samples); DISPLAY_X=0; DISPLAY_Y=0; originalX=0; originalY=0; PIXEL_RATIO_X=1.0; PIXEL_RATIO_Y=1.0; READY=false; ObjectsDeleteAll(ChartID(),system_tag); EventSetMillisecondTimer(44); ResourceFree("TESTRESOURCE"); return(INIT_SUCCEEDED); } void OnTimer(){ EventKillTimer(); //load if(ResourceCreate("TESTRESOURCE","\\Files\\"+testBMP)){ uint pixels[],width,height; if(ResourceReadImage("::TESTRESOURCE",pixels,width,height)){ //collect samples int total_samples=0; uint capture=ColorToARGB(fillColor,255); ArrayResize(samples,width*height,0); //scan the image originalX=(int)width; originalY=(int)height; int co=-1; for(int y=0;y<(int)height;y++){ for(int x=0;x<(int)width;x++){ co++; if(pixels[co]==capture){ total_samples++; samples[total_samples-1].x=x; samples[total_samples-1].y=y; samples[total_samples-1].xd=x; samples[total_samples-1].yd=y; } } } ArrayResize(samples,total_samples,0); Print("Found "+IntegerToString(total_samples)+" samples"); //--------------------------------------------transformation tests //we have 2 features OR 2 dimensions //first blind step is covariance matrix of all features /* so what happens here ? We fill up a matrix where : each row is a sample each column is a feature */ matrix original; original.Init(total_samples,2); //fill for(int i=0;i<total_samples;i++){ original[i][0]=samples[i].xd; original[i][1]=samples[i].yd; } //lets do 1 , no loop matrix covariance_matrix=original.Cov(false); Print("Covariance matrix"); Print(covariance_matrix); matrix eigenvectors; vector eigenvalues; matrix inverse_covariance_matrix=covariance_matrix.Inv(); if(covariance_matrix.Eig(eigenvectors,eigenvalues)){ Print("Eigenvectors"); Print(eigenvectors); Print("Eigenvalues"); Print(eigenvalues); //apply for(int i=0;i<total_samples;i++){ vector thissample={samples[i].xd,samples[i].yd}; thissample=thissample.MatMul(eigenvectors); samples[i].xd=thissample[0]; samples[i].yd=thissample[1]; if(RemoveCovariance){ samples[i].xd/=MathSqrt(eigenvalues[0]); samples[i].yd/=MathSqrt(eigenvalues[1]); } /* samples[i].xd*=eigenvalues[0]; samples[i].xd/=(eigenvalues[0]+eigenvalues[1]); samples[i].yd*=eigenvalues[1]; samples[i].yd/=(eigenvalues[0]+eigenvalues[1]); */ } //reconstruct double minX=INT_MAX,maxX=INT_MIN,minY=INT_MAX,maxY=INT_MIN; for(int i=0;i<total_samples;i++){ if(samples[i].xd>maxX){maxX=samples[i].xd;} if(samples[i].xd<minX){minX=samples[i].xd;} if(samples[i].yd>maxY){maxY=samples[i].yd;} if(samples[i].yd<minY){minY=samples[i].yd;} } //0 to 1 double rangeX=maxX-minX; double rangeY=maxY-minY; double allMax=MathMax(maxX,maxY); double allMin=MathMin(minX,minY); double allRange=allMax-allMin; for(int i=0;i<total_samples;i++){ samples[i].xd=((samples[i].xd-minX)/rangeX)*1000.0; samples[i].yd=((samples[i].yd-minY)/rangeY)*1000.0; /* samples[i].xd=((samples[i].xd-allMin)/allRange)*1000.0; samples[i].yd=((samples[i].yd-allMin)/allRange)*1000.0; */ samples[i].x=(int)samples[i].xd; samples[i].y=(int)samples[i].yd; } originalX=1000; originalY=1000; }else{Print("Cannot eigen");} //--------------------------------------------transformation tests build_deck(originalX,originalY); READY=true; }else{ Print("Cannot read image"); } }else{ Print("Cannot load file"); } ExpertRemove(); Print("DONE"); } void build_deck(int img_x, int img_y){ //calculation of display size int screen_x=(int)ChartGetInteger(ChartID(),CHART_WIDTH_IN_PIXELS,0); int screen_y=(int)ChartGetInteger(ChartID(),CHART_HEIGHT_IN_PIXELS,0); //btn size int btn_height=40; screen_y-=btn_height; //fit double img_x_by_y=((double)img_x)/((double)img_y); //a. to width int test_x=screen_x; int test_y=(int)(((double)test_x)/img_x_by_y); if(test_y>screen_y){ test_y=screen_y; test_x=(int)(((double)test_y)*img_x_by_y); } //placement int px=(screen_x-test_x)/2; int py=(screen_y-test_y)/2; DISPLAY.CreateBitmapLabel(ChartID(),0,system_tag+"_display",px,py,test_x,test_y,COLOR_FORMAT_ARGB_NORMALIZE); DISPLAY_X=test_x; DISPLAY_Y=test_y; DISPLAY.Erase(0); PIXEL_RATIO_X=((double)(DISPLAY_X))/((double)(img_x)); PIXEL_RATIO_Y=((double)(DISPLAY_Y))/((double)(img_y)); PIXEL_RATIO_X=MathMax(1.0,PIXEL_RATIO_X); PIXEL_RATIO_Y=MathMax(1.0,PIXEL_RATIO_Y); //override PIXEL_RATIO_X=8.0; PIXEL_RATIO_Y=8.0; update_deck(); } void update_deck(){ DISPLAY.Erase(ColorToARGB(clrBlack,255)); uint BASECOLOR=ColorToARGB(BaseColor,255); //sdraw for(int i=0;i<ArraySize(samples);i++){ double newx=(((double)samples[i].x)/((double)originalX))*((double)DISPLAY_X); double newy=(((double)samples[i].y)/((double)originalY))*((double)DISPLAY_Y); int x1=(int)MathFloor(newx-PIXEL_RATIO_X/2.00); int x2=(int)MathFloor(newx+PIXEL_RATIO_X/2.00); int y1=(int)MathFloor(newy-PIXEL_RATIO_Y/2.00); int y2=(int)MathFloor(newy+PIXEL_RATIO_Y/2.00); DISPLAY.FillRectangle(x1,y1,x2,y2,BASECOLOR); } DISPLAY.Update(true); ChartRedraw(); } void OnDeinit(const int reason) { } void OnTick() { }
Edit : Found an article on how to do the feature composition :
Although i think , not sure , but i think you don't reduce from 2 features to 1 feature . The wording is wrong.
You create one feature from 2 features and you are left with one new feature.
So if we had 100 features and we did the following we'd not get 99 features but one feature , one new feature.
I think.